Contact Us
CiiS Lab
Johns Hopkins University
112 Hackerman Hall
3400 N. Charles Street
Baltimore, MD 21218
Directions
Lab Director
Russell Taylor
127 Hackerman Hall
rht@jhu.edu
Last updated: Apr 26th, 2019
Standard X-ray imaging brings difficulty for surgeons to identify region of interest (ROI) features from anatomical clutter. Dual energy X-ray enables anatomical clutter reduction via material decomposition by utilizing the physical properties of X-ray formulations. The figure below illustrates the decomposition process. Traditional Dual Energy X-ray Absorptiometry (DEXA) system has been developed in analyzing bone density, fat tissue, etc. But the model is largely approximated and simplified, because the target ROI is usually large and not targeted to small region accuracy. Thus, we propose to re-design the algorithm and improve the accuracy on decomposition of injected cement during femoroplasty. Decomposed frames can be used to improve 3D reconstruction of the injected cement, to better understand the cement distribution.
Conventional X-ray imaging is not sufficient to characterize object precisely, especially in the aspect of density, material identity, volume thickness, 3D depth of the object, etc. It is then hard for the surgeons to identify Region of Interest(ROI) using X-rays with multiple material stacked intensities. Thus, there is a need to develop an efficient decomposition system that can separate multiple materials in projection domain.
This project targets at utilizing the physical formulation of X-ray including the material attenuation coefficient, material density, X-ray casting length and especially photon spectrum energy. Here is how this formula looks like,
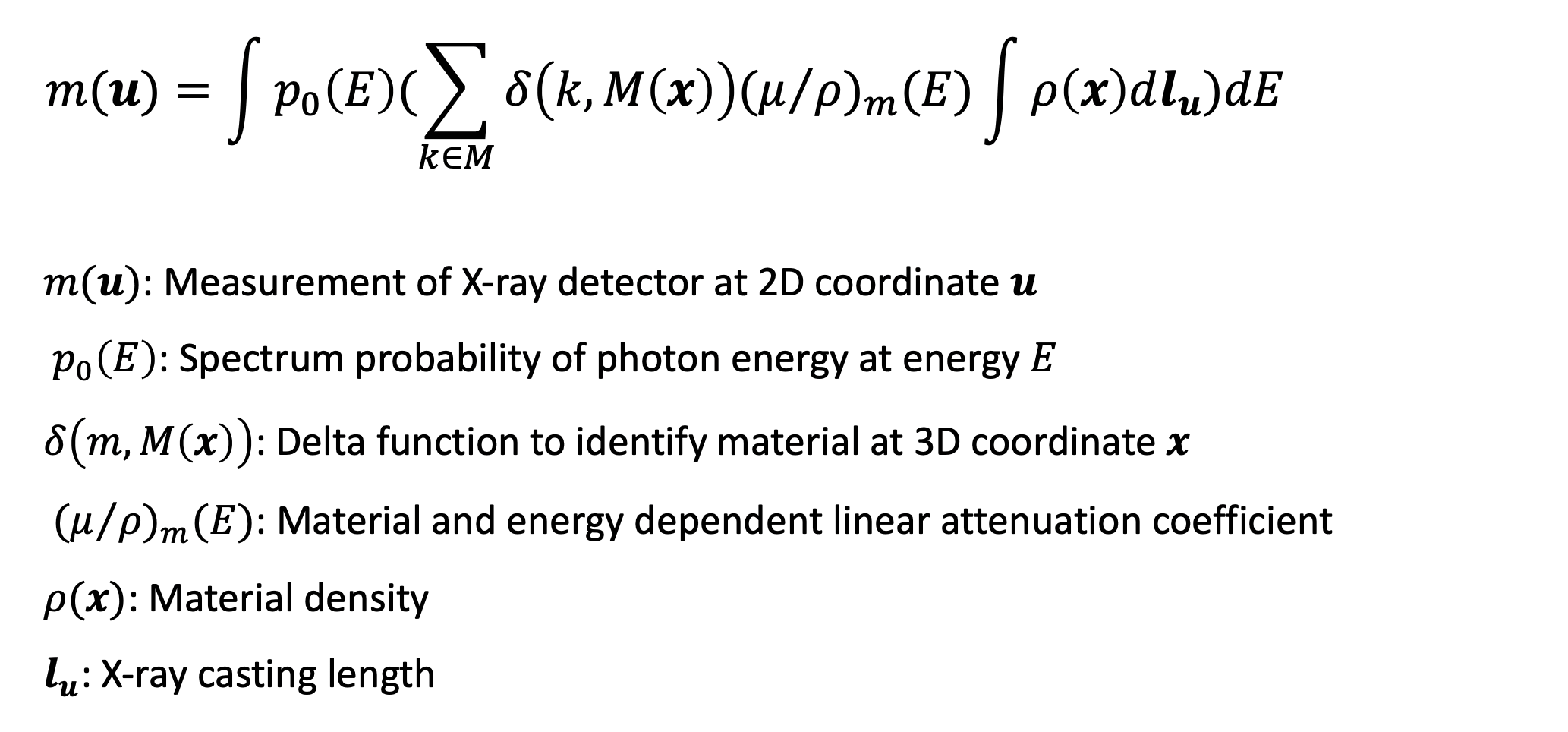
First, we conducted analysis using traditional model-based method. In the case of two materials, we can then further simplify this formula by its parameter w and the desired unknowns T for each pixel u:
The photon spectrum can be simulated using the online SIEMENS opensource simulation software. Below is the simulation result for 60kV and 75kV stop energy spectrum
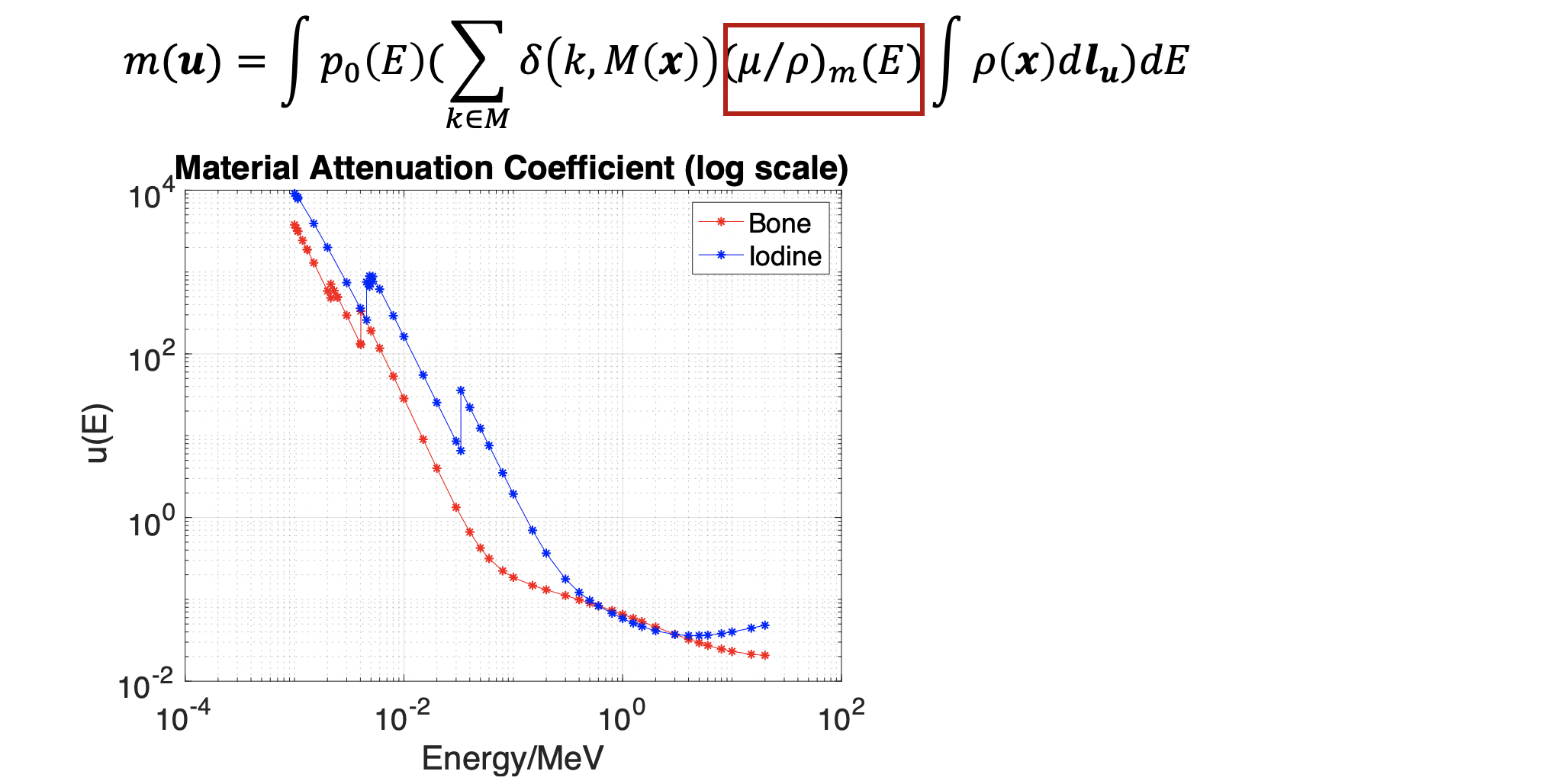
The material attenuation coefficient is also simulated using the NDE open source data.(https://www.nde-ed.org/EducationResources/CommunityCollege/Radiography/Physics/attenuationCoef.htm) We can see an obvious K-edge effect (coefficient jump as particular energy level).
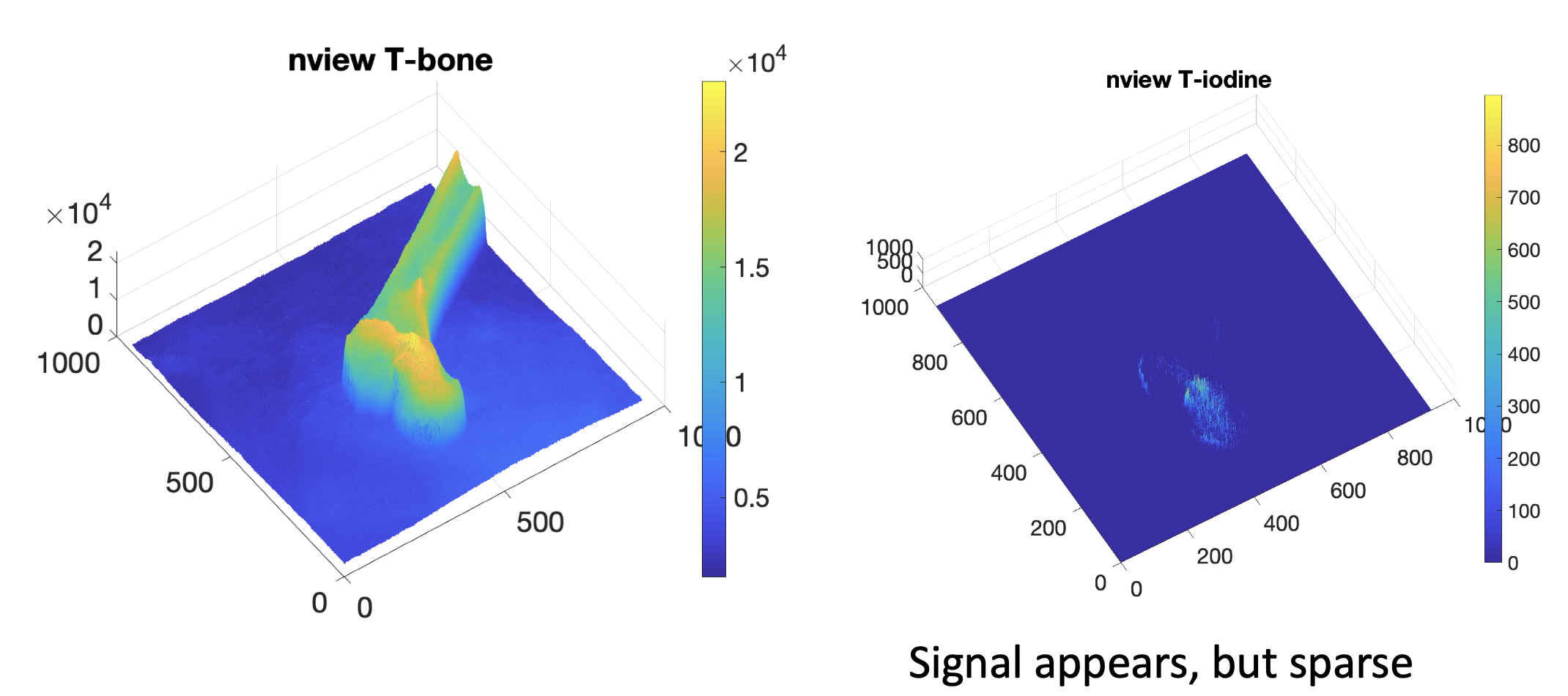
For Real X-ray validation, we plan to use the nView system, which is a fast 3D reconstruction system using low dose X-ray projections. Because the X-ray projections and reconstruction have correspondence with this system, its 3D reconstruction data can be used to label material groundtruth for validation. We plan to use this nView system to conduct femoroplasty injection experiment and collect dual-energy dataset during cement injection process for validation of the proposed algorithm.
Below is the figure of real X-ray acquisition:
Preliminary Result
By solving the linear model, we get the least square solution, but because this model is non-constrained, the output is highly overfitted to this data, which leads to negative T(u) problem, violating the physical constraints. Then we added a non-negative constraint to this least square solution. Below is the output,
In order to conduct simulation study, we want to use the recently proposed X-ray simulation framework – DeepDRR, which can perform fast and realistic simulation of fluoroscopy and digital radiography from CT scans by using deep learning. We can also simulate X-ray projections with different energy levels. Another benefit by introducing DeepDRR is that it enables segmentation in 3D domain, including bone, soft tissue and air, which can be used to generate target decomposition projections as groundtruth images for training.
Simulation study will start from testing on two materials. Iodine is our first target, because it has an obvious K-edge jump in its photon attenuation at around 70keV (shown in Figure. 3), which is a very good feature to do dual-energy decomposition. We plan to first simulate bone injection cement inside the femur, which mainly composes of iodine, and try to decompose it from the other background materials. If it works well, we then plan to test on more complicated situations. The right figure is the CT data we are using for simulation.
Simulation Results
Here are some simulation results of DeepDRR from a pelvis CT scan. The high energy image is the original simulation. The soft tissue and bone are generated from the traditional method.
We propose to introduce deep learning to this prediction framework, by explicitly including the physical constraint in the estimation part. Figure presents the overall workflow of the proposed pipeline. The input is the dual-energy acquisitions. We first run the traditional constrained least square solution to get an initialization result T(u). Then we try to train a discriminator to model the probability of the current solution P(T|M). This trained model is then used as a loss function to solve the overall combined optimization objective. This method will include both model driven and learning based method, which will avoid overfitting to training data, but biased by the physical model.
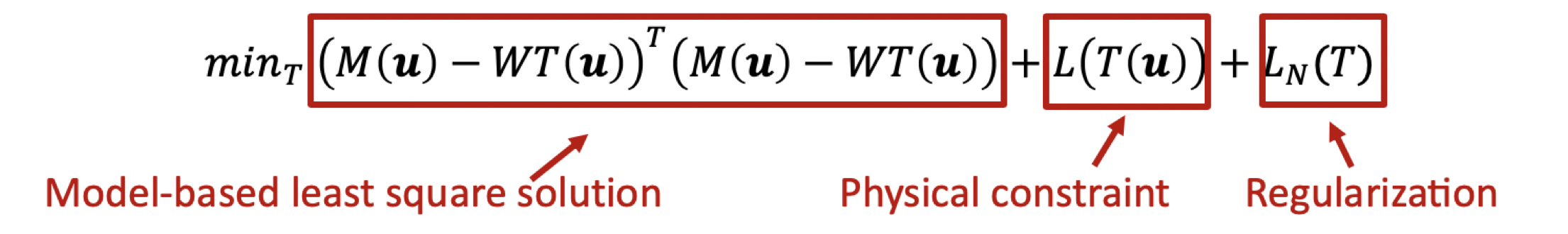
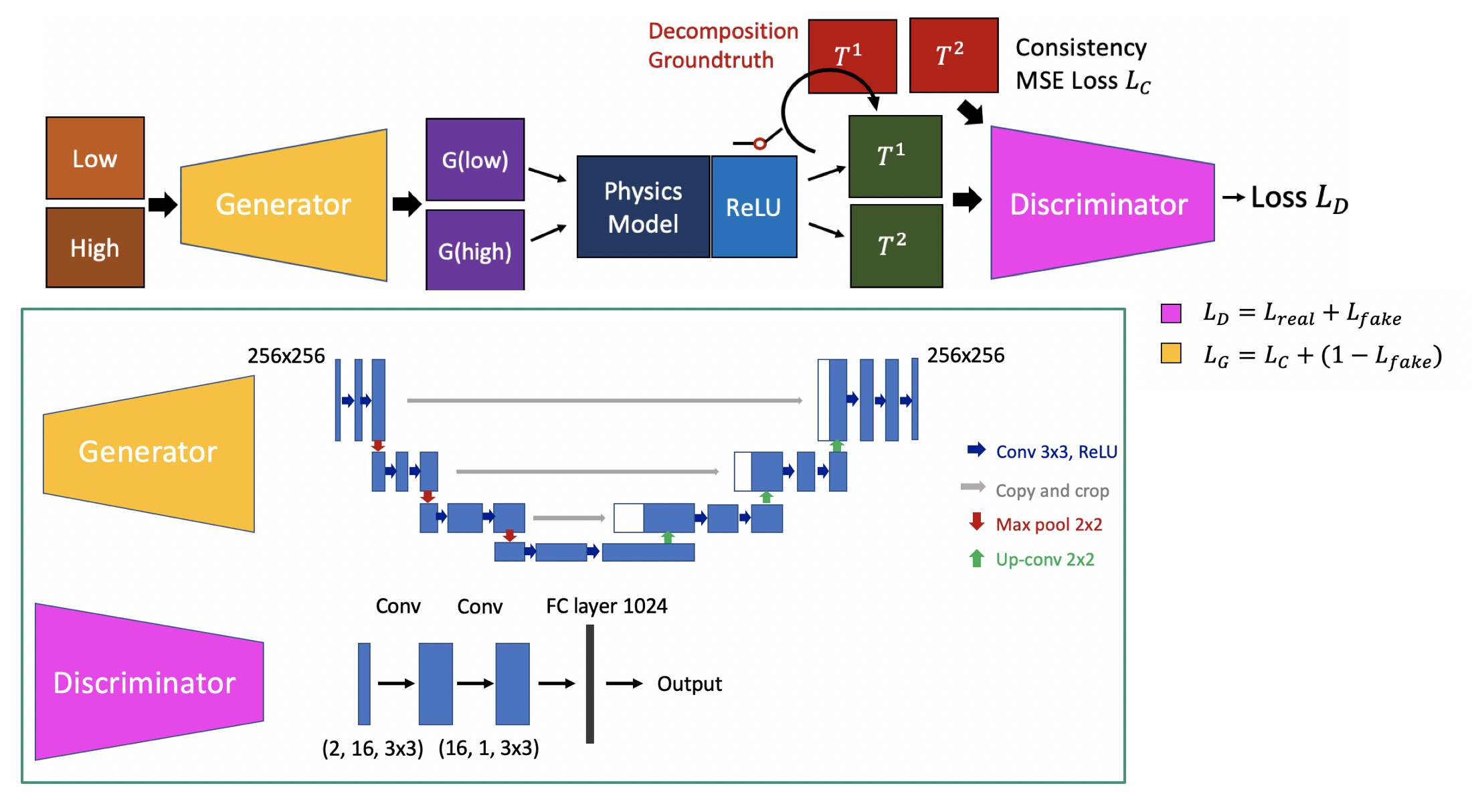
We first tested the results on the synthetic validation dataset. The accuracy is reported as mean dice score compared with the groundtruth decomposition and the signal variation. Since the predictions all have background noise, we are using a threshold of 1.2 times mean response to filter the noise. Then we compared the iodine prediction mask with the groundtruth mask. The mean dice score for model based method is 0.268, while our learning based method reaches 0.791. We also calculated the mean standard deviation of the output iodine response. The model based method has mean STD of 0.0037, while our learning based method is 0.0028. So the learning based method is largely outperforming model based method in prediction accuracy and smoothness of response.
We then tested the model on the unseen real X-rays. The constrained least squared solution has very sparse and limited signals, which are mostly be- cause of the overfitting of the linear model. Our learning-based model yields much better result in the cement region. Fig. 8 presents the synthetic valida- tion result and the real X-ray cement decomposition result. We can clearly see that the model based version has stronger noise and bad background prediction in the bone region, while the learning based method has more smooth signals and flatten background, which is closer to the real solution. The real X-ray decomposition also shows the shape of the injection tunnel and the strong distribution at the center of the head, which corresponds the real experiment condition.
Here give list of other project files (e.g., source code) associated with the project. If these are online give a link to an appropriate external repository or to uploaded media files under this name space (2019-99).