Table of Contents
Semi-Automated Brain Tumor Segmentation
Summary
In this project, we aim to apply theoretical improvements to the watershed transformation to MRI images of glioblastoma patients. This assisted segmentation tool aims to increase accuracy and reduce inter and intra-observer variability present in current segmentation practices. A C++ implementation of this algorithm will be developed within the Insight Toolkit (ITK) library.
- Students: Alexander Liu, Nathaniel Tippens
- Mentor(s): Dr. Hadie Adams, Dr. Alfredo Quiñones-Hinojosa
Background, Specific Aims, and Significance
The segmentation of brain tumor MRI scans is of critical importance in the evaluation of tumor progression. To evaluate progression of a tumor and the subsequent treatment response, the most commonly used methods to determine treatment responses in brain tumors are the Macdonald criteria and the Response Evaluation Criteria in Solid Tumors (RECIST) criteria. The Macdonald criteria incorporates two-dimensional measurements with steroid dosing and the patients’ neurological examinations, while the RECIST criteria evaluates tumor response based on measurement of the longest one-dimensional (1D) diameter. Both these criterions only consider two dimensions, and are ultimately rough estimates of a brain tumor’s shape and features.
Our specific aims are:
- To promote the use of volumetric analysis in brain tumors, which provides a more accurate quantification of tumor burden.
- To create freely available, easy-to-use software that decreases segmentation time and reduces inter- and intra-observer variability, allowing for rapid determination of tumor volume.
Deliverables
- Minimum:
- Implement a 2D watershed algorithm in ITK
- Expected:
- Implement 2D watershed algorithm in ITK
- Integrate algorithm into ITK-SNAP
- Test variability and accuracy of the program
- Perform segmentations on simulated datasets
- Maximum:
- Implement 3D watershed algorithm in ITK
- Integrate algorithm into ITK-SNAP
- Test variability and accuracy of the program
- Perform segmentations on simulated datasets
- Investigate inter/intra-observer variability
Technical Approach
There are no published segmentation methods that have been validated on the full variety of high grade gliomas. To ensure accurate segmentation of even the most difficult gliomas, we have chosen a semi-automated, or user-assisted, segmentation mode. Semi-automated methods include active contouring, intensity thresholding, level-set segmentation, and watershed segmentation. We have chosen to develop an interactive watershed-based segmentation method because it generates significant partitions of an image and relies on the user for high-level interpretation of each of these regions. The algorithm is very fast, and translates to a simple point-and-click interface where what you see is what you get. Additionally, the segmentation is always the same, which we hope will greatly reduce both intra- and inter-operator variability. Besides reducing variability with the watershed approach, we can also increase the speed of segmentation compared to manual segmentation, since the user no longer needs to trace borders by hand. Instead, the user is merely choosing regions that they consider to be part of a lesion. We hope to apply recent improvements to the watershed transformation that have not yet been used for medical image segmentation. Additionally, we will evaluate the software's performance (meaning accuracy and variability) on both simulated datasets (where absolute tumor volume is known) and real datasets, using multiple trained observers.
Milestones and Progress
- Watershed Implementation
- Planned Date: 3/10/11
- Expected Date: 3/10/11
- Status: Complete
- Viscous Watershed Implementation
- Planned Date: 4/14/11
- Expected Date: 4/14/11
- Status: Coded & Compiled. Executables and Source uploaded.
- Journal Submission and Code Documentation
- Planned Date: 5/13/11
- Expected Date: 5/13/11
- Status: Journal Submission in Preparation, Code Documentation finished
Major changes or issues
- RESOLVED: Linking issue with previously implemented ComponentTree class.
- Variability and accuracy study deferred until usable program is developed.
Results
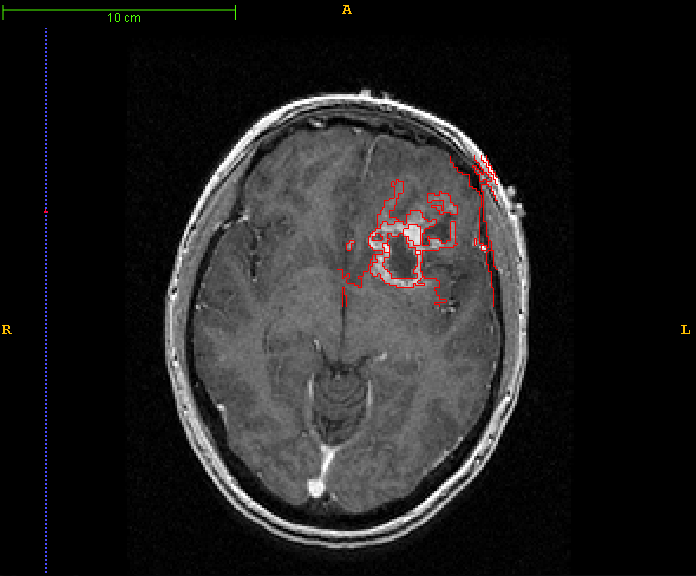
- Leaky regions for some tumor scans with poorly defined borders with unmodified watershed algorithm.
- Viscous watershed algorithm finished. Necrosis merging with gray matter resolved, at the cost of enhancement rings.
- Algorithm Specification & Documentation Finished
Reports and Presentations
- Project Plan
- Project Background Reading
- Project Checkpoint
- Project Final Presentation
Project Bibliography
- Kanaly CW, Ding D, Mehta AI, Waller AF, Crocker I, et al. 2011 A Novel Method for Volumetric MRI Response Assessment of Enhancing Brain Tumors. PLoS ONE 6(1): e16031. doi:10.1371/journal.pone.0016031
- Marloes M.J. Letteboer, Ole F. Olsen, Erik B. Dam, Peter W.A. Willems, Max A. Viergever, Wiro J. Niessen, Segmentation of Tumors in Magnetic Resonance Brain Images Using an Interactive Multiscale Watershed Algorithm, Academic Radiology, Volume 11, Issue 10, October 2004, Pages 1125-1138, ISSN 1076-6332, DOI: 10.1016/j.acra.2004.05.020.
- Cates, J.E., Whitaker, R.T. & Jones, G.M. Case study: an evaluation of user-assisted hierarchical watershed segmentation. Medical Image Analysis 9, 566-578 (2005).
- Fiez, J. A., H. Damasio, et al. (2000). “Lesion segmentation and manual warping to a reference brain: Intra- and interobserver reliability.” Human Brain Mapping 9(4): 192-211.
- Mazzara, G.P., Velthuizen, R.P., Pearlman, J.L., Greenberg, H.M. & Wagner, H. Brain tumor target volume determination for radiation treatment planning through automated MRI segmentation. International Journal of Radiation Oncology*Biology*Physics 59, 300-312 (2004).
- Renz, D., et al. Accuracy and reproducibility of a novel semi-automatic segmentation technique for MR volumetry of the pituitary gland. Neuroradiology, 1-12 (2010).
- Marcel Prastawa, Elizabeth Bullitt, Guido Gerig, Simulation of brain tumors in MR images for evaluation of segmentation efficacy, Medical Image Analysis, Volume 13, Issue 2, Includes Special Section on Functional Imaging and Modelling of the Heart, April 2009, Pages 297-311, ISSN 1361-8415, DOI: 10.1016/j.media.2008.11.002.
- Yushkevich, P. A., J. Piven, et al. (2006). “User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability.” NeuroImage 31(3): 1116-1128.
- Vachier, C. and F. Meyer (2005). “The Viscous Watershed Transform.” Journal of Mathematical Imaging and Vision 22(2): 251-267.
- Cousty, J., G. Bertrand, et al. (2009). “Watershed Cuts: Minimum Spanning Forests and the Drop of Water Principle.” Pattern Analysis and Machine Intelligence, IEEE Transactions on 31(8): 1362-1374.
- Letteboer, M. M. J., O. F. Olsen, et al. (2004). “Segmentation of tumors in magnetic resonance brain images using an interactive multiscale watershed algorithm.” Academic Radiology 11(10): 1125-1138.
- Sergio E. Hernandez, Kenneth E. Barner and Yu Yuan, “Region merging using homogeneity and edge integrity for watershed-based image segmentation”, Opt. Eng. 44, 017004 (Dec. 21, 2004); doi:10.1117/1.1830042