Table of Contents
Intraoperative Registration of Pathology for Adjuvant Postoperative Radiotherapy
Last updated: May 8, 2014; 8:40 PM
Summary
Team Members:
• Kareem Fakhoury
• Matthew Hauser
• Steven Lin
Mentors:
• Dr. Harry Quon, MD
• Dr. Junghoon Lee, PhD
• Dr. Jeremy Richmon, MD
Background, Specific Aims, and Significance
With regards to cancers in the head and neck, the general procedure for eliminating the pathology is to perform surgical resection of the tumor as well as pre- and post-operative radiotherapy. Adjuvant radiotherapy is delivered based on a plan created after the surgery based on pre-operative and post-operative CT scans as well as reports from the operating surgeon. However, post-operative tissue deformation – shifts in the anatomy surrounding the surgical area – makes the previous location of the tumor difficult to identify. Because of this uncertainty and in order to ensure that none of the remaining cancer cells are missed, the area identified for radiotherapy is overestimated. This is harmful to patients because the volume of irradiated tissue dictates the toxicity affecting the patient, which has negative downstream consequences, such as intense pain and the inability to swallow and eat. The goal of our project is to show how the tissue around the surgical area deforms from pre- to post-operative CT scans. This will allow radiation oncologists and dosimetrists to more accurately localize the area containing the remaining cancer cells. This information, in turn, will inform planning to allow tighter and more accurate volumes for adjuvant radiotherapy. Small decreases in irradiated volume will lead to significant decreases in toxicity.
Deliverables
- Minimum: (Expected by March 15 - Completed March 15)
Data collection and comparison of deformable registration algorithms. The data collection is a rather lengthy process due to the many factors that are involved; such as acquiring the pig-heads, and coordinating a time where Dr. Richmond and technicians would be available. Comparison of the existing deformable registration algorithms may be a lengthy process despite utilizing currently available libraries.
- Expected: (Expected by April 4)
Minimum deliverables and implementation of workflow to register image data and evaluate tissue deformation. The ultimate goal for identifying points of pathology using the Polaris requires further implementation. This may result in a change of workflow in surgery. After data collection and algorithms comparison, we will work with Dr. Richmond to devise a feasible alteration to the current surgery workflow.
- Maximum: (Expected by mid-May)
Expected deliverables, determining optimal marker placements for tissue deformation evaluation and writing a journal paper. The marker points will be used to identify the deformation of the tissue around the wound. Currently, Dr. Richmond will place three points around the wound at his discretion. We hope to identify the optimal distance and number of points in relation to the size of the wound and expected tissue deformation. If time permits, we will submit/ publish a journal article about our findings.
- We also added a new maximum deliverable of determining the feasibility of registration of the tongue outside the mouth, but due to time constraints we are focusing on the more realistic situation of the tongue being inside the mouth.
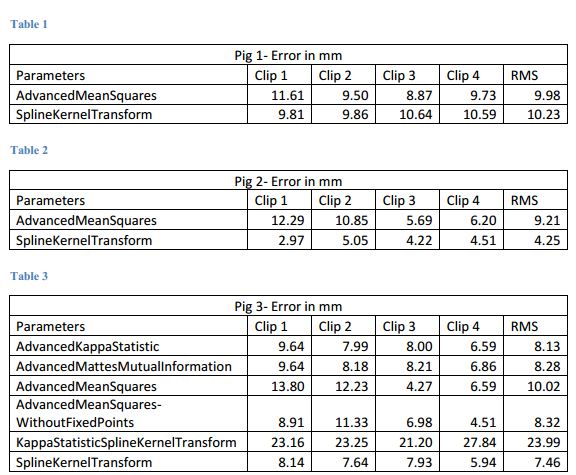
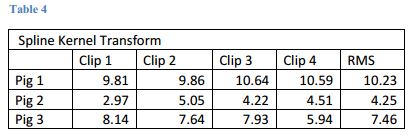
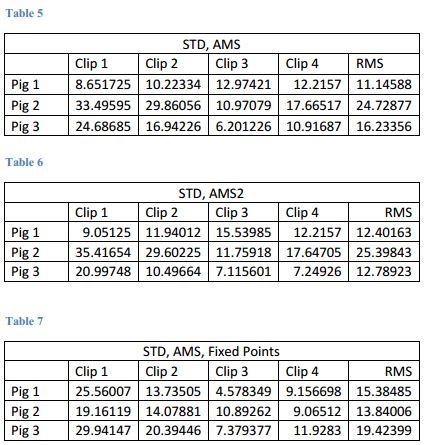
- These are our results
- Comparison of various parameter methods
- Comparison of all using the Spline Kernel Transform
- Comparison of various parameter files when registering the closed image to the preop image and ultimately to open image
Technical Approach
First, we will acquire the pre-operative, intra-operative and post-operative data. These will be obtained with the help Dr. Richmond and Dr. Quon. Currently, we plan to use pig-heads because of they have similar structures to humans. The following is a brief methodology to obtain the data:
1. CT scan of the three pig-heads before the operation
2. Tissue removal from tongue in each pig head
3. Marking around the site of removal with the Polaris marker
4. Surgical clips will be placed where the Polaris has marked.
5. CT scan of each head with an wound still open
6. Close the wound
7. CT scan of each head with a closed wound
The first step to developing the registration algorithm is to compare and evaluate current deformable registration algorithms. It is possible that there are algorithms that would be effective at registering the pre-operative to post-operative. There are many algorithms on the subject as well as open-source libraries that would help to make the testing phase more efficient. The intra-operative Polaris data and the post-operative surgical clips location will allow us to measure the accuracy of each algorithm. After this initial research phase, we will identify and develop an algorithm that will be appropriate. It is likely that a currently available algorithm is appropriate and would require minor changes to fit this operation.
Dependencies
Data collection - Experiment will be conducted at JHH using pig heads.
i. 4 pig heads – Will be purchased at Mt. Airy Locker Co. One pig head will be put through a CT head in order to determine whether pig heads are appropriate for this experiment.
- First pig head obtained. Three other pig heads needed for next data collection day, TBD.
ii. CT scans - Department of Radiological Oncology and will be coordinated by Dr. Quon with the help of Wen Liu.
- Arrangements successfully made by Dr. Quon.
iii. Lab space reservation - Coordinated by Dr. Quon
- Arrangements successfully made by Dr. Quon.
iv. Polaris tracking system – At the generosity of Dr. Taylor
- Polaris obtained for first experiment with relative ease. Will need for next data collection day, TBD.
All funding provided by Dr. Quon
Milestones and Status
• March 10 (changed from March 3 due to technical difficulties): This is our planned preliminary Data Collection Day, during which 1 pig head will have cookie-cutter resection, and Polaris data as well as pre- and post-operative images will be collected. We also plan to understand how to use Polaris and its data by this date. Members responsible: Kareem and Matt.
- Completed March 10. See figure 1.
• March 7: Complete literature review by this date. Members responsible: all.
- Completed, various dates for each member but all completed by March 7
• March 15: We plan to finish evaluating and comparing all open-source deformable registration algorithms of interest by this date. Members responsible: all.
- Completed March 13.
• March 15: Devise method to incorporate Polaris data into pre-operative data by this date: Members responsible: Steven and Kareem.
- Completed March 14. See figures 2 and 3.
• April 7 (orig. March 21): Implement registration of pre- to post-operative CT data by this date. Members responsible: Matt and Steven.
- Determined input parameters to optimize registration. See figures 4 and 5.
- April 9: We completed the second data collection experiment in which we used three pig heads. We will use these images to improve our registration method.
- May 4: We have made much progress with image registrations. We are still tweaking the parameters to make the registrations as accurate as possible so that we can see how closely the tongue markers move compared to where the gold standard clips are.
- May 8: We have analyzed data to determine extent to which tongue markers outlining the pathology accurately move with the movement of the tongue. Results are far from perfect, but they serve as a guideline as to how well the pathology can be localized and as a stepping stone to further work.
• April 11 (orig. April 4): We plan to have the polished workflow finished by this date. Members responsible: all.
- May 4: The workflow is complete, but we are still tweaking the parameters used in the image registrations. Otherwise, all other pieces of the workflow are in order.
- Completed May 8 with registration parameter files.
• April 16 (orig. April 15): Once the implementation is complete, we plan to find optimal tracker placement by running the data collection with varied placement. Members responsible: all.
- We will not be able to achieve this deliverable due to time constraints and the great amount of time and effort required for data collection in the CT Sim room with the pig heads.
• Mid-May: We plan to submit a paper for publication. Members responsible: all.
- Rather than submitting a paper for publication we will help Dr. Quon with his grant submission.
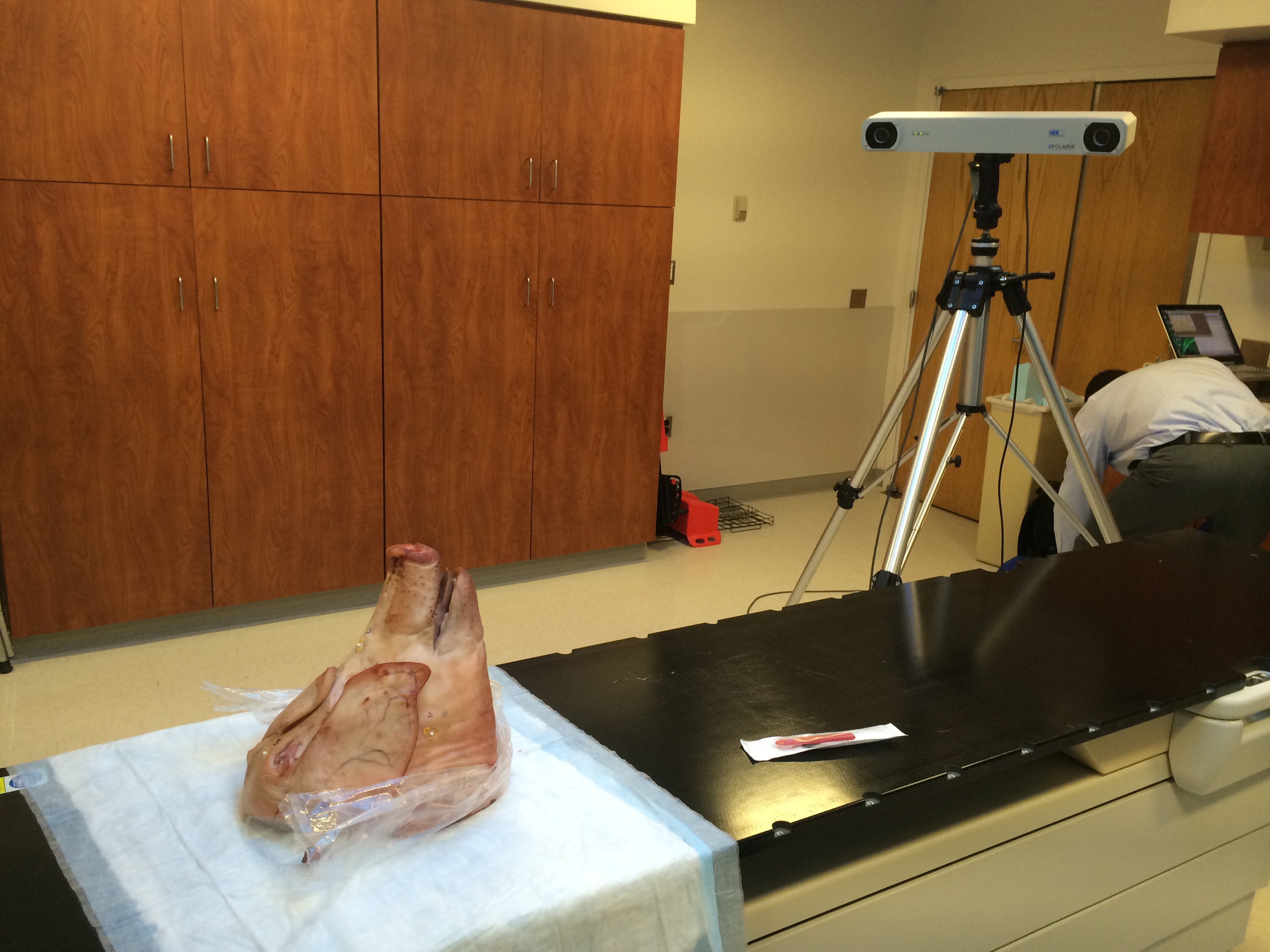 Figure 1. Data collection set up.
Figure 1. Data collection set up.
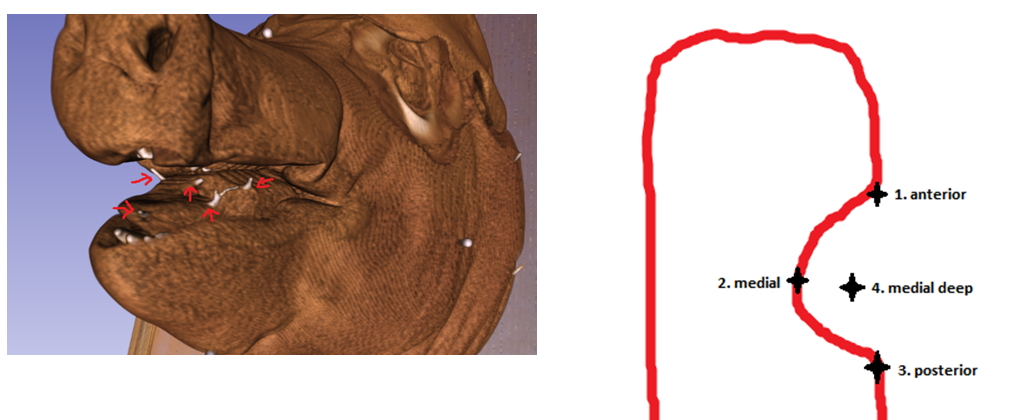
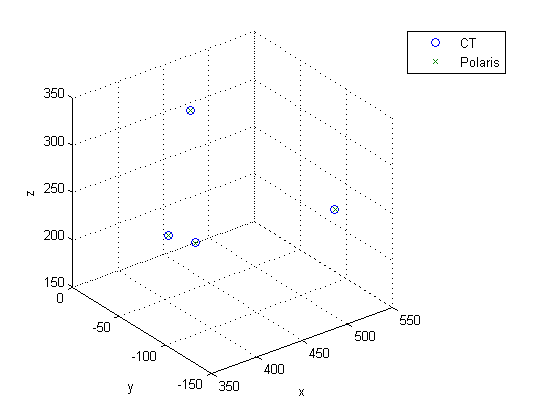 Figure 2. Shows how registered polaris points outlining pathology line up with the gold standard surgical clips as they appear in the CT scan.
Figure 2. Shows how registered polaris points outlining pathology line up with the gold standard surgical clips as they appear in the CT scan.
 Figure 3. The blue circle indicates a polaris point added to the CT scan.
Figure 3. The blue circle indicates a polaris point added to the CT scan.
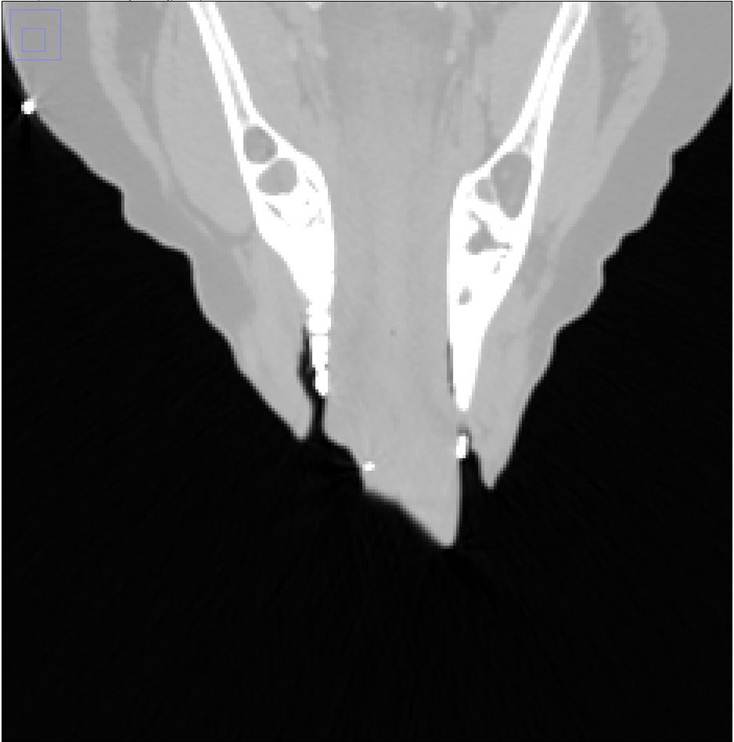
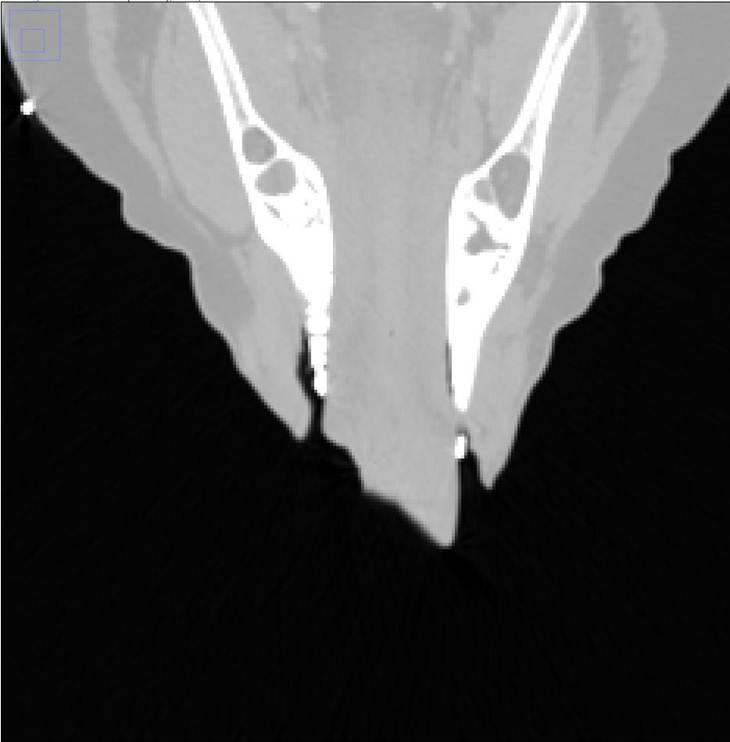 Figure 4. The image on the left is the original with the clips showing up white. These were removed by thresholding, shown on the right.
Figure 4. The image on the left is the original with the clips showing up white. These were removed by thresholding, shown on the right.
 Figure 5. Registration results showing registration of preoperative image to postoperative open wound image.
Figure 5. Registration results showing registration of preoperative image to postoperative open wound image.
Reports and presentations
- Project Plan
- Project Background Reading
- See Bibliography below for links.
- Project Checkpoint
- Paper Seminar Presentations
- Project Final Presentation
- Project Final Report
- links to any appendices or other material
Project Bibliography
S. Klein, M. Staring, K. Murphy, M. A. Viergever, and J. P. W. Pluim, “elastix: A toolbox for intensity-based medical image registration,” IEEE Transactions on Medical Imaging, Vol. 29, No. 1, pp.196-205, 2010.
B. B. Avants, C. L. Epstein, M. Grossman, and J. C. Gee, “Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegerative brain,” Medical Image Analysis, Vol. 12, No. 1, pp.26-41, 2008. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2276735/
H. Lester and S. R. Arridge. A survey of hierarchical non-linear medical image registration. Pattern Recognit., 32(1):129 – 149, 1999.