Table of Contents
Assessing Ventilator-Associated Pneumonia (VAP) in the PICU
Last Updated: 5/09/2019
Summary
Our objective is to determine if we can identify VAP risk early through analyzing time series data and other identification markers gathered in the PICU. There is a multidisciplinary team working on addressing this issue; we are specifically working with the radiology team to focus on X-ray image changes associated with VAP risk. Our goal is to develop an algorithm that can accurately predict if a patient has VAP by analyzing time-series images throughout a patient’s addressed history.
- Students: Suraj Shah
- Mentor(s): Drs. Mathias Unberath, Jim Fackler, and Jules Bergmann
Background, Specific Aims, and Significance
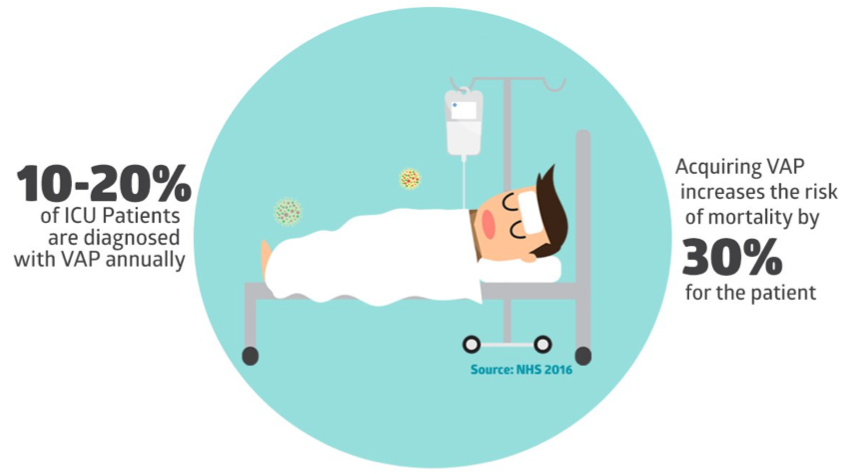
Clinical Background: Ventilator-associated Pneumonia (VAP), “specifically refers to pneumonia developing in a mechanically ventilated patient more than 48 hours after tracheal intubation.”(1) When under ventilation, patients are in a critical, life-sustaining ICU therapy; the body is at a fragile state and susceptible to diseases, including bacterial infections that are attributed to VAP. There are other risks involved as well, as further disease progression, volume overload, latrogenic infection, and ventilator injury. VAP has had such a deteriorating effect in the ICU ward that it is now the leading cause of mortality among nosocomial infections and the leading cause of nosocomial morbidity (1). Specifically, acquiring VAP increases the risk of morbidity by 30% for any given patient. This is compounded by the fact that 10-20% of ICU patients are diagnosed with VAP annually. Ventilator-associated complications are also correlated with a much greater length of stay and time under ventilation. This leads to greater strain on the entire healthcare value chain, from the provider, insurer, and most importantly, the patient.
Motivation and significance: While there is a multi-disciplinary team working on addressing the issues arising from VAP in the ICU (including the PICU team working on identifying biomarkers and the ID team working on appropriate cultures and antibiotics), there hasn’t been a comprehensive study connecting the radiology component of monitoring patients with VAP and/or risk of VAP. This is mainly due to the clinical data collection process. The X-ray images that are collected occur over many different hospitals, at different orientations of patients, on different machines, and either at inspiry or expiry. There is not a standardized process for data collection, thus leading to an aggregation of thousands of X-ray images, but ones that are not easily ingestible into an algorithm that is able to readily classify the risk of VAP for a specific patient. For example, the largest chest X-ray dataset of adult images, MIMICS CXR, contains over 224,000 images of over 60,000 patients in various studies. Furthermore, when applying an algorithm to pediatric patients, we must understand that the patients grow much more quickly over time, thus any algorithm must account for the changes in image dimension.
Therefore, there is need for a protocol development that can aggregate X-ray information and an algorithm that can accurately help to predict occurrence of VAP based image-related features. This specifically manifests itself in the following:
- Aggregating the largest available public datasets to provide the most complete picture of chest X-ray information (start with adult-only images)
- Training several neural net models on a static image to reach state-of-art levels of pneumonia prediction
- Identifying the features most relevant in a physician's diagnosis of pneumonia through saliency mapping and class activation
- Produce clustering methods on top of pre-trained networks to output unsupervised learning which can be applicable in any setting
We hope our project will lead to a renewed focus in the ICU on these high-risk patients, while avoiding unnecessary therapy for low-risk patients.
Deliverables
- Minimum: (Expected by April 14th)
- A database of X-ray cohorts and segment them based off patient type, thoracic pathologies, and other characteristics [COMPLETED]
- Trained algorithm (pytorch model) for static image prediction [COMPLETED]
- Expected:
- Trained algorithm (pytorch model) for multi-class classification [COMPLETED]
- Trained saliency mapping algorithm for physician use [COMPLETED]
- Maximum: (Expected by May 9th)
- A trained pytorch model file for unsupervised learning of visual features
- Clinically actionable report of F1 scoring compared against current radiologists
Technical Approach
The project workflow is split into the following main components:
- Screening and Collecting Working Data to Build Database of X-Ray Images
- Assembling an Input/Output Module for Analyzing Images within the Database
- Performing Saliency Mapping and Class Activation on Image Features
- Using Unsupervised Learning Methods to Cluster Images
The bulk of our time will be focused on the second component, testing models and determining the best-performing neural network to classify VAP occurrence on a static image.
Data Aggregation and Screening
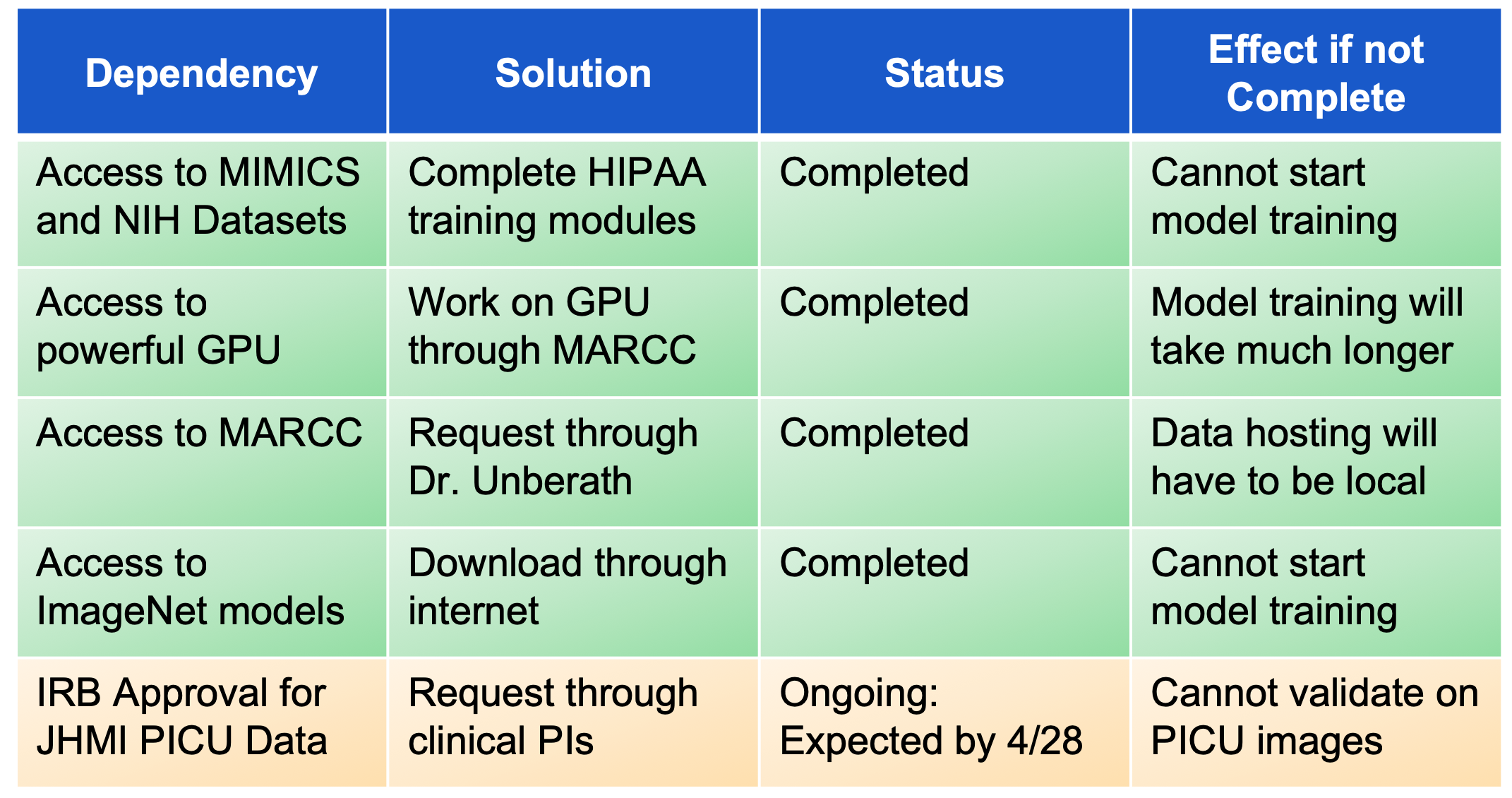
First, we will be assembling a database of chest X-Ray images which will be hosted through MARCC (Maryland Advanced Research Computing Center). This database will comprise of data collected from publicly available datasets such as MIMICS CXR, NIH, etc. We are focusing first on the publicly available datasets as we await IRB approval for the pediatric data from JHU. These datasets will be screened for abnormalities and outliers and cleaned accordingly so. Initially, we believed that we had access to time-series data of X-ray images; while this is not the case, there is still a wide scope of work we can accomplish with classification techniques.
Assembling Input/Output Module – Convolutional Neural Network Classification
To first get a pulse for how our system will analyze chest X-ray images, we will need to create a static image predictor – namely being, choosing an apt neural network to correctly classify a patient’s occurrence of VAP based off one image. Once we can train and identify the best performing neural net for static image prediction, we will be able to use the time-series images for a more accurate prognosis. Potential neural networks to consider include:
- VGG-16: developed in 2014, this network consists of 16 layers, and is appealing because of the uniform architecture. In terms of performance, it is one of the most preferred choices extracting features from images, which could be very relevant when pursuing relevant factors in the chest X-ray images. One concern with VGG is that has 138 million parameters, which could be a potential hurdle in terms of difficulty to handle.
- ResNet: also known as Residual Neural Network, this was developed in 2015 and focuses heavily on on “skip connections” (gated recurrent units used in RNNs) and batch normalization. The architecture consists of 152 layers but is much less parameter dependent, thus reducing the complexity involved.
- DenseNet: an extension of ResNet. DenseNet’s methodology proposes concatenating outputs from the previous layers instead of using the summation (ResNet merges previous layers with future leaders).
The main goal here is to reduce the number of links between layers for more efficient and accurate processing.
Before ingesting the data into the training set pipeline, we will be analyzing each neural net and the strength of each (based off image properties). We will start by training the networks on a specified training dataset and subsequently testing it on specified training set. After determining a set accuracy threshold with our mentors and clinical collaborators, we can identify the best performing neural net. We will have to fine-tune the parameters of the neural net to best apply to our data, and perhaps split the data more rigorously (i.e. omitting multiple copies of the same patient) so that we are able to have the best performing model possible.
Performing Saliency Mapping and Class Activation on Image Features In computer vision, a saliency map is an image that shows each pixel's unique quality. The goal of a saliency map is to simplify and/or change the representation of an image into something that is more meaningful and easier to analyze. This is highly important when applying our model to the Chest X-ray images. Beyond prediction alone, it is extremely helpful for physicians (and can help them be more efficient) to highlight where exactly the source of pneumonia (or any other thoracic pathology) is on a certain image. Previous work has been done to identify regions of interest for the physician. Thus, our aim is to apply saliency mapping frameworks to highlight the regions on the Ches X-ray images after our model has correctly identified a case of pneumonia.
Using Unsupervised Learning Methods to Cluster Images The most highly-used convolutional neural networks are trained on ImageNet, which contains 1 million images to cover most types of classification (up to 1000 labels). To increase performance, one could feasibly increase the size of the dataset by a factor of 10-100x, but that would concern much more manual annotation, placing a burden on human effort and is much more extensive than what is currently available in the data science community. Thus, it is imperative to produce a model that can create generalizations for visual features to apply to any large-scale dataset that does not require supervision. This is critical to understanding of classification for my project because of the complexities of X-ray data when applied to pneumonia diagnoses with different demographics (specifically ages), ventilations, and machines. One goal of our collaborators is to be able to apply the data in the PICU – however, most of the publicly available data has only adult chest X-ray data. Applying unsupervised clustering methods might unlock generalization of pneumonia features in my project that previously was not available with supervised learning. Thus, for my maximum deliverable, I will be applying the DeepCluster technique developed my Mathilde et. al. to cluster the features produced by the CNN. The insight gathered here is focused on clustering features instead of direct labels, which could be potentially more important in thoracic pathology classification.
Dependencies
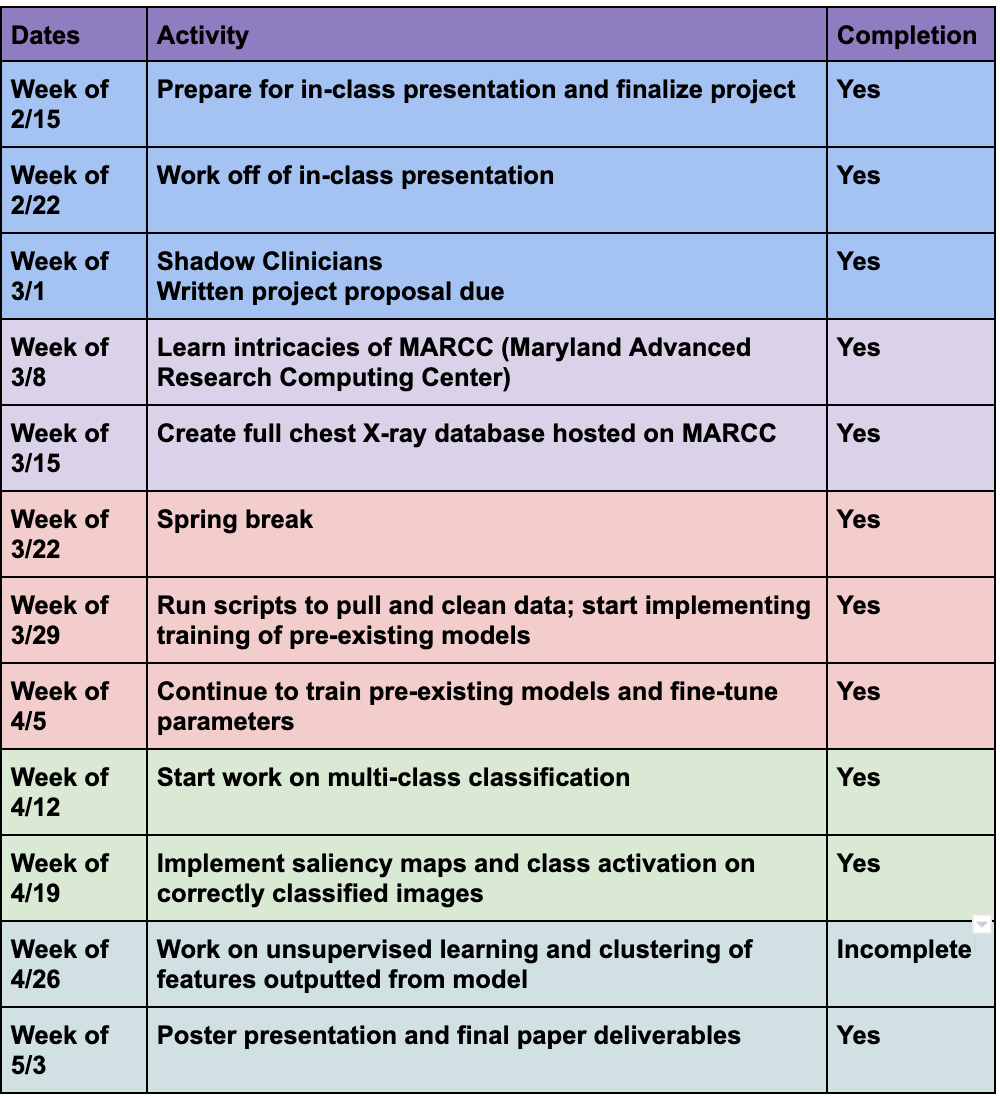
Milestones and Status
Reports and presentations
- Project Plan
- Project Background Reading
- See Bibliography below for links.
- Project Checkpoint
- Paper Seminar Presentations
- Project Final Presentation
- Project Slide Teaser
- Project Final Report
- Technical Appendices
Project Bibliography
- Kollef, M. H., Dr. (2005). WHAT IS VENTILATOR-ASSOCIATED PNEUMONIA AND WHY IS IT IMPORTANT? Respiratory Care, 50(6), 714-724. Retrieved February 26, 2019
- Chen, Y., Pont-Tuset, J., Montes, A. and Van Gool, L. (2019). Blazingly Fast Video Object Segmentation with Pixel-Wise Metric Learning. [online] arXiv.org. Available at: https://arxiv.org/abs/1804.03131 [Accessed 18 Feb. 2019].
- Yin Y, Hoffman EA, Ding K, Reinhardt JM, Lin CL. A cubic B-spline-based hybrid registration of lung CT images for a dynamic airway geometric model with large deformation. Phys Med Biol. 2010;56(1):203-18.
- He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770-778.
- He, K., Zhang, X., Ren, S. and Sun, J. (2019). Identity Mappings in Deep Residual Networks. [online] arXiv.org. Available at: https://arxiv.org/abs/1603.05027 [Accessed 18 Feb. 2019].
- Hatami, N., Gavet, Y. and Debayle, J. (2019). Classification of Time-Series Images Using Deep Convolutional Neural Networks. [online] arXiv.org. Available at: https://arxiv.org/abs/1710.00886 [Accessed 20 Feb. 2019].
- Mathilde Caron, Piotr Bojanowski, Armand Joulin, and Matthijs Douze. “Deep Clustering for Unsupervised Learning of Visual Features.” arXiv:1807.05520 [cs.CV]. Proc. ECCV (2018).
- “Pneumonia Can Be Prevented-Vaccines Can Help | CDC.” Centers for Disease Control and Prevention. 2018. Centers for Disease Control and Prevention. 20 Apr. 2019 <https://www.cdc.gov/pneumonia/prevention.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Ffeatures%2Fpneumonia%2Findex.html>.
- Kirton, Orlando. “Mechanical Ventilation - The American Association for the Surgery of Trauma.” American Association for the Surgery of Trauma, AAST, 2011. 20 Apr. 2019. www.aast.org/GeneralInformation/mechanicalventilation.aspx.
- Johnson AEW, Pollard TJ, Berkowitz S, Greenbaum NR, Lungren MP, Deng C-Y, Mark RG, Horng S. MIMIC-CXR: A large publicly available database of labeled chest radiographs. arXiv (2019).
- Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng C-K, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 101(23):e215-e220 [Circulation Electronic Pages; http://circ.ahajournals.org/content/101/23/e215.full]; 2000 (June 13).
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. IEEE CVPR 2017, http://openaccess.thecvf.com/content_cvpr_2017/papers/Wang_ChestX-ray8_Hospital-Scale_Chest_CVPR_2017_paper.pdf (link is external)
- Karen Simonyan, Andrew Zisserman. “Very Deep Convolutional Networks For Large-Scale Image Recognition.” arXiv:1409.1556v6 [cs.CV] 10 Apr 2015
- Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun. “Deep Residual Learning for Image Recognition.” arXiv:1512.03385 [cs.CV] 10 Dec 2015
- Huang, Gao and Liu, Zhuang and van der Maaten, Laurens and Weinberger, Kilian. “Densely connected convolutional networks.” arXiv:1608.06993 [cs.CV]. Proc. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017)
- Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, Antonio Torralba. “Learning Deep Features for Discriminative Localization.” Computer Science and Artificial Intelligence Laboratory, MIT. Proc. CVPR (2016)
Other Resources and Project Files
[[https://github.com/surajshah980/CISIIProject] Technical Appendix and Source Code]