Table of Contents
Project Name
Last updated: May 10
Summary
The creation of an automatic segmentation method for use in CT spine based upon the Max-Flow/Min-Cut algorithm.
- Students: Niko Eng, Ben Ramsay
- Mentor(s): Dr. Jeff Siewerdsen and Dr. Tharindu De Silva
Background, Specific Aims, and Significance
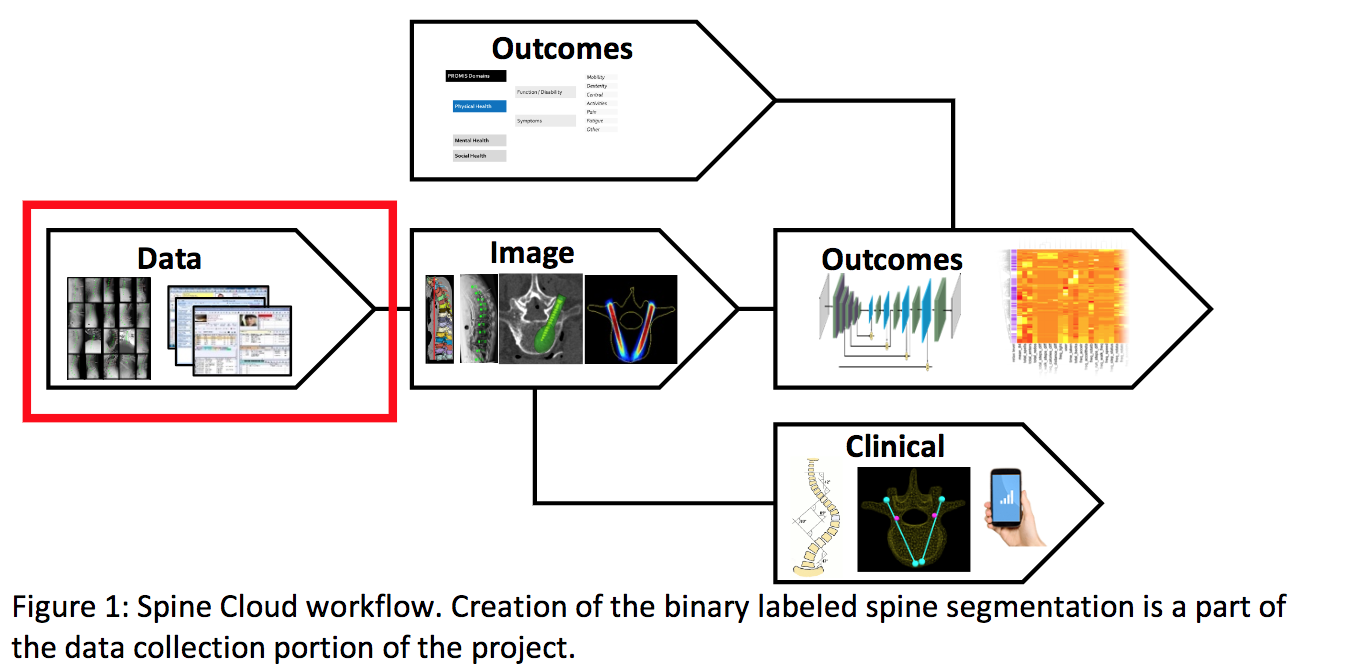
The overall goal of our project is to develop a state-of-the-art automatic segmentation method for spine CT images using max-flow/min-cut optimization. We are motivated for this specific task, since our work will be a major component of “Spine Cloud” a multi-year project proposed by the Dr. Siewerdsen’s I-STAR lab. “Spine Cloud” hopes to curate a database consisting of patient demographic data, image and specific anatomy, surgical procedures, and pathologies. Once organized, we hope to correlate these defined clinical variables and automatic image analysis to patient surgical outcomes. By developing this highly quantitative approach on how to approach future spine surgeries, “Spine Cloud” will provide more favorable and consistent outcomes.
A necessary component of “Spine Cloud” is a large database of annotated spine CT images based on accurate, automatic segmentation. Currently within the I-STAR lab, segmentation of spine CTs is handed manually which while accurate is often time-consuming. While there are simple techniques for auto-segmentation like Thresholding and Region Growing that are computationally efficient and easy to implement, they often fail to give accurate segmentation. With these techniques, each voxel is treated independent in the CT image, which prohibits local neighborhood relations like smoothness and curvature to be accounted for during segmentation. Instead, we propose to treat segmentation as an energy minimization problem which can account for local relations by transforming the CT image into a graph and using Max-Flow / Min-Cut Optimization in order to minimize the energy function.
Original Deliverables
- Minimum: (Expected by March 9th)
Access accuracy of the N20 manually segmented validation datasetImplementation of the Max-Flow/Min-Cut segmentation algorithm for the spine
- Expected: (Expected by April 20th)
Analysis of parameter sensitivityEvaluation of segmentation accuracyGeneration of a large N200 dataset for SpineCloud
- Maximum: (Expected by May 15th)
Method for patient-specific parameter selectionMethod to accommodate spine anomalies
Updated Deliverables
- Minimum: (Expected by March 9th) Completed
- Access accuracy of the N20 manually segmented validation dataset
- Implementation of 3D Max-Flow/Min-Cut segmentation algorithm for the spine
- Expected: (Expected by May 6th) In Progress
- Analysis of parameter sensitivity on N20 (Completed)
- Generate and Evaluate Segmentation Accuracy on N20 (Completed)
- Automate Intensity Profiling for N200 (Completed)
- Manual Segmentation of Patients in N200 for Validation (In Progress)
- Maximum: (Expected by May 15th) To Be Done
- Accurate Segmentation of N200 Lumbar CT (without Spinous Process) (In Progress)
- Address anomalies in N200 (spine instrumentation, cancer)
Technical Approach
Our segmentation relies on the implementation of a 2D max-flow min-cut optimization method. The MATLAB code to execute this optimization was given to us by our mentors. Upon receiving the code we explored the parameter space to see what affects certain parameters would have on the segmentation. To access the accuracy of our segmentation we implemented root mean squared error(RMSE) and dice coefficient metrics. Our implementation was tested using these metrics on the manually segmented N20 spine CT dataset. Once we had a good understanding of the algorithm, we shifted our method from a 2D implementation to a 3D implementation. We also incorporated information of centroid positions into our algorithm which informed us of the center of each vertebrae in the CT image. Once our methodology was working in the N20, our mentors encouraged us to continue to adjust the algorithm to segment the N200 dataset.
Since the N200 is not manually segmented, the only way to evaluate the performance is qualitatively. For this reason, our mentors encouraged us to segment one of the N200 members so that our segmentation method could be validated in the N200. Our approach to the N200 dataset was to first segment the spine without the spinous process, and then attempt to incorporate the spinous process and address certain anomalies like instrumentation and tumors that might throw off the segmentation. The segmented N200 dataset will be directly used as a data repository for the Spine Cloud project.
Background of Graph Cuts
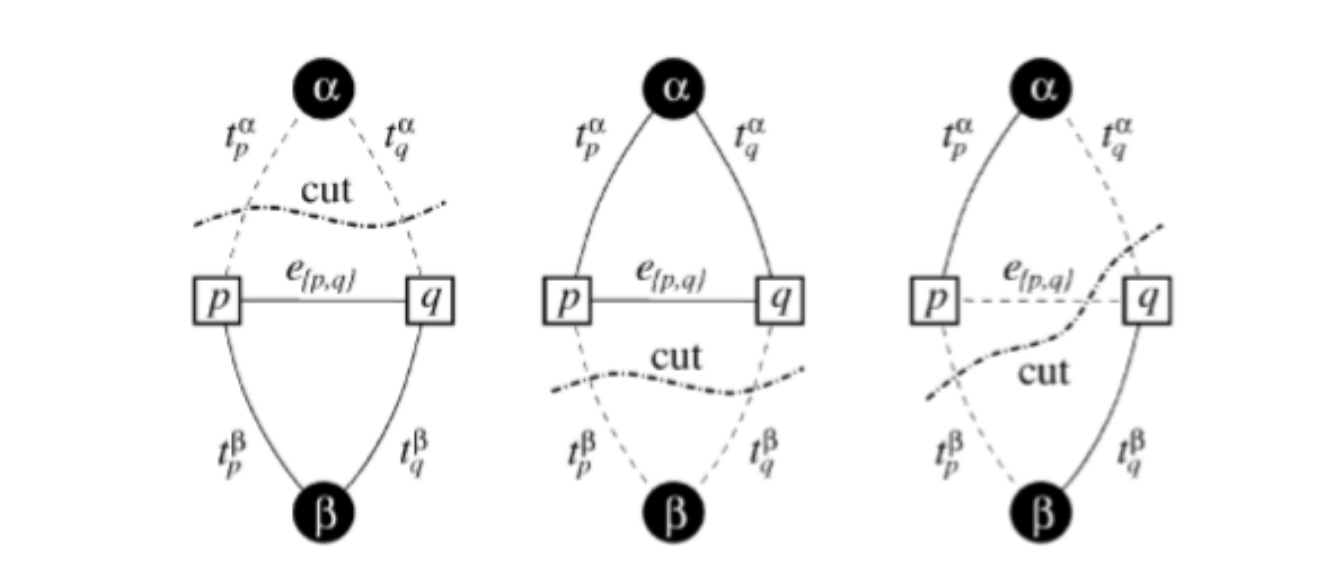
For a weighted graph with two vertices and α,β terminals, the graph cut C is a set of edges that separates the two terminals in the induced graph such that no flow can go from one terminal node to the other. The cost of cut C is the sum of the edge weights. Therefore, the minimum cut is the cheapest cut possible that separates the two terminals. This cut represents the Min-Cut / Max-Flow algorithm.
Figure B1: Examples of Different Graph Cuts Separating the two Terminals α, β
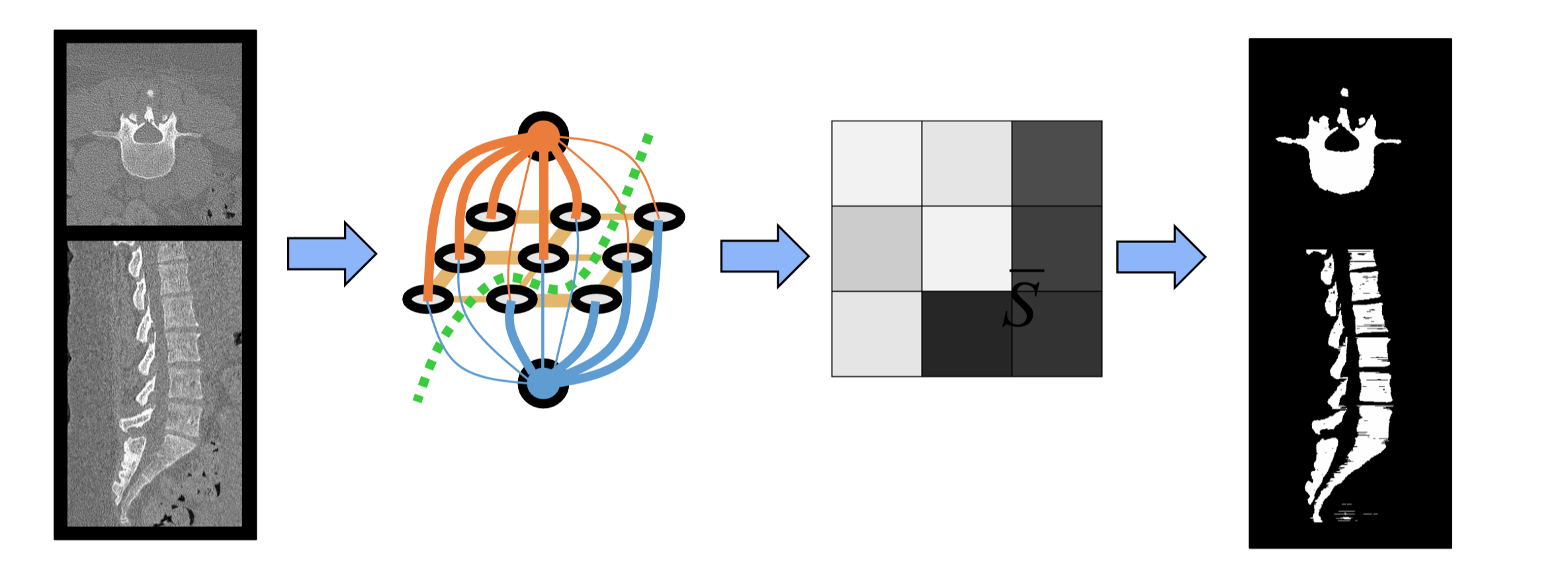
As an example of how our method applies graph-cuts, we first start with a CT image of the spine (Figure B2). To define the nodes of the graph, we take each voxel in the CT image and create a node (gray) and then define two terminal nodes (orange and blue). Each voxel node is connected by an edge to every neighboring voxel node as well as to both terminal nodes.
Figure B2: Example of Graph-Cut Segmentation for Spine CTs
The values of the terminal link weights are based on prior information detailing the likelihood that certain voxel are a part of the spine or the background. These priors ideally will have different likelihoods for spine vs background allowing for the best separation between bone and background. Voxel node to voxel node edges are determined by measurable quantities of the image and contribute to how smooth the segmentation will appear. Ultimately, the edge weights determine the final cut that will be made since the cut made must minimize the sum of the weights which are cut. In this minimization, the terminal nodes are separated from each other and the resulting segmentation is assigned based on which terminal node a given voxel is connect to.
Original Dependencies
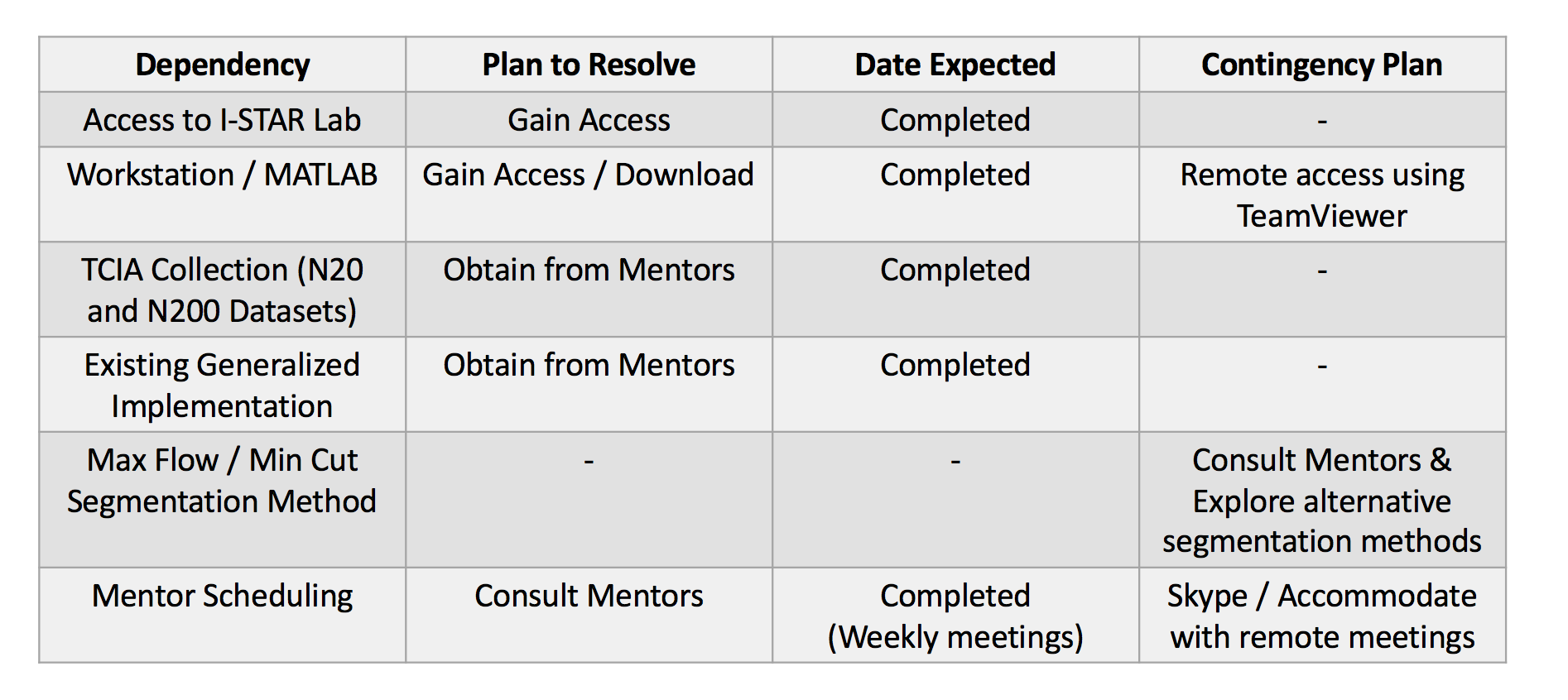
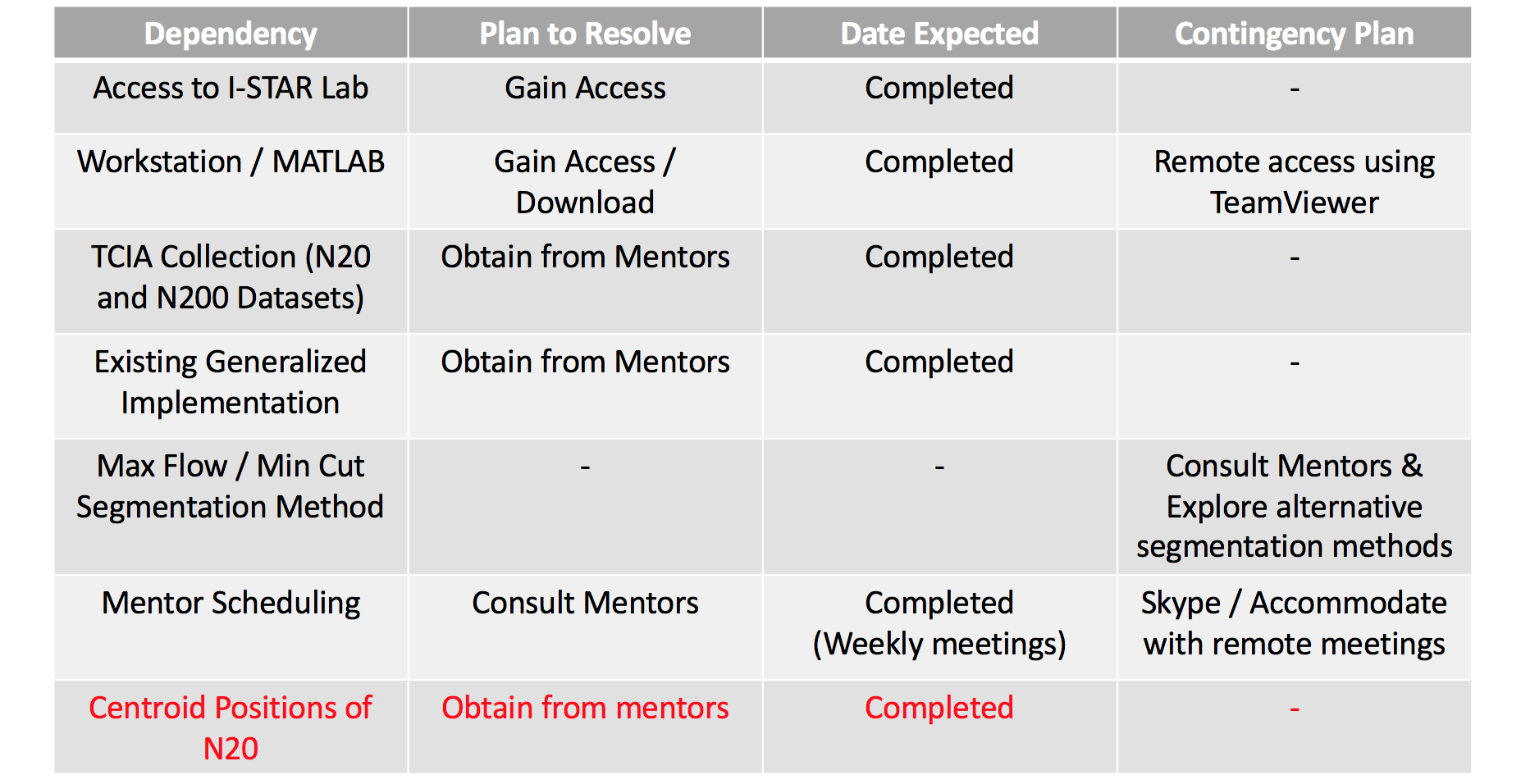
Updated Dependencies
Milestones and Status
- Milestone 1: Implementation of the 2D Max-Flow/Min-Cut segmentation algorithm for the spine
- Planned Date: March 9th
- Expected Date: March 9th
- Status: Completed
- Milestone 2: Analyze Basic Parameter Sensitivity on Single N20 Case
- Planned Date: March 23rd
- Expected Date: March 23rd
- Status: Completed
- Milestone 3: Implementation of quantitative accuracy metrics (RMSE & Dice)
- Planned Date: March 30th
- Expected Date: March 30th
- Status: Completed
- Milestone 4: Use centroid to create axial cylindrical distance weighting
- Planned Date: March 30th
- Expected Date: March 30th
- Status: Completed
- Milestone 5: Implementation of the 3D Max-Flow/Min-Cut segmentation algorithm for the spine
- Planned Date: April 6th
- Expected Date: April 6th
- Status: Completed
- Milestone 6: Create non-axial cylindrical distance weighting
- Planned Date: April 6th
- Expected Date: April 6th
- Status: Completed
- Milestone 7: Identify intervertebral disc space using non-axial convolutional cylinder
- Planned Date: April 13th
- Expected Date: April 13th
- Status: Completed
- Milestone 8: Analyze Basic Parameter Sensitivity on entire N20
- Planned Date: April 18th
- Expected Date: April 18th
- Status: Completed
- Milestone 9: Apply Distance Weighting for N200
- Planned Date: April 27th
- Expected Date: April 27th
- Status: Completed
- Milestone 10: Automate Intensity Profiling for N200
- Planned Date: May 8th
- Expected Date: May 8th
- Status: Completed
- Milestone 12: Manually Segment Patients in the N200 dataset for quantitative Validation
- Planned Date: May 15th
- Expected Date: May 15th
- Status: Working
- Milestone 13: Accurately segment the N200 lumbar without spinous process
- Planned Date: May 15th
- Expected Date: May 15th
- Status: Working
- Milestone 14: Accommodate abnormalities in the N200 dataset
- Planned Date: May 15th
- Expected Date: May 15th
- Status: Working
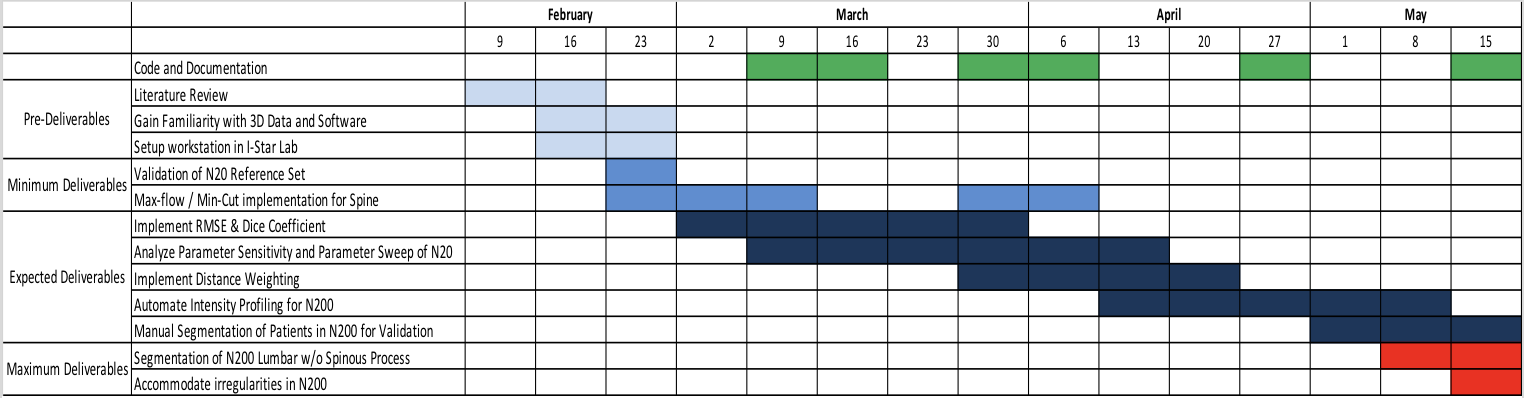
Project Timeline
Reports and presentations
- Project Plan
- Project Background Reading
- See Bibliography below for links.
- Project Checkpoint
- Paper Seminar Presentations
- Project Final Poster
- Project Final Report
- links to any appendices or other material
Project Bibliography
- Yuan, Jing, et al. “A Study on Continuous Max-Flow and Min-Cut Approaches.” 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2010
- Boykov, Y.y., and M.-P. Jolly. “Interactive Graph Cuts for Optimal Boundary & Region Segmentation of Objects in N-D Images.” Proceedings Eighth IEEE International Conference on Computer Vision. ICCV 2001
- Boykov, Yuri, and Vladimir Kolmogorov. “An Experimental Comparison of Min-Cut/Max-Flow Algorithms for Energy Minimization in Vision.” Lecture Notes in Computer Science Energy Minimization Methods in Computer Vision and Pattern Recognition, 2001, pp. 359–374.
Other Resources and Project Files
Here give list of other project files (e.g., source code) associated with the project. If these are online give a link to an appropriate external repository or to uploaded media files under this name space.2018-21