Contact Us
CiiS Lab
Johns Hopkins University
112 Hackerman Hall
3400 N. Charles Street
Baltimore, MD 21218
Directions
Lab Director
Russell Taylor
127 Hackerman Hall
rht@jhu.edu
One common theme in research in our laboratory and elsewhere is combinining prior information about an individual patient or patient populations with new images to produce an improved model of the patient.
Examples may be found throughout our research writeups(e.g., Sparse X-Ray Reconstruction, 3D ACL Tunnel Position Estimation). Some more examples may be found below.
Just as other groups, we have a long history of creating statistical models of patient anatomy
and using the results for such purposes as assisting the segmentation of 3D images by means of deformable
registration of these models to patient-specific images (e.g., [Yao 2003a], [Ellingsen 2009],
[Chintalapani 2007], [Chintalapani 2009], [Seshamani 2011])
and to create estimates of 3D models from sparse sets of 2D data (e.g., [Yao 2002a], [Yao 2003b],
[Sadowsky 2006], [Sadowsky 2007], [Chintalapani 2009b], [Chintalapani 2010]).
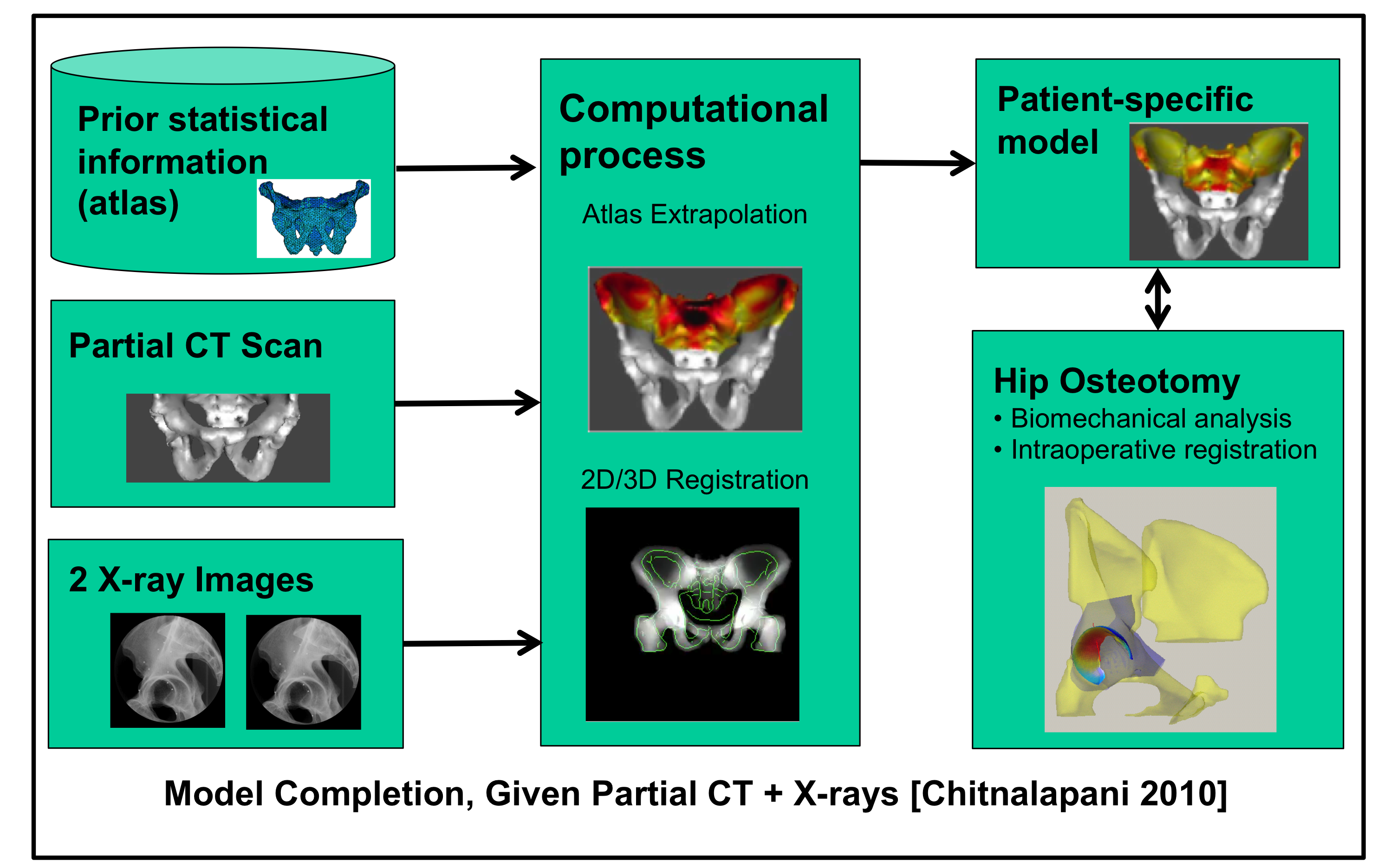 Information may be fused from multiple sources to produce a model. For example, Chintalapani et al.
have combined preoperative CT images of a limited portion of the pelvis with a statistical atlas and
a limited number of x-ray images to create a model that may be used for preoperative planning
and intraoperative registration in periacetabular osteotomies [Chintalapani 2010].
A fuller description of the overall system may be found in [Arminger 2007] and [Lepisto 2008].
Information may be fused from multiple sources to produce a model. For example, Chintalapani et al.
have combined preoperative CT images of a limited portion of the pelvis with a statistical atlas and
a limited number of x-ray images to create a model that may be used for preoperative planning
and intraoperative registration in periacetabular osteotomies [Chintalapani 2010].
A fuller description of the overall system may be found in [Arminger 2007] and [Lepisto 2008].
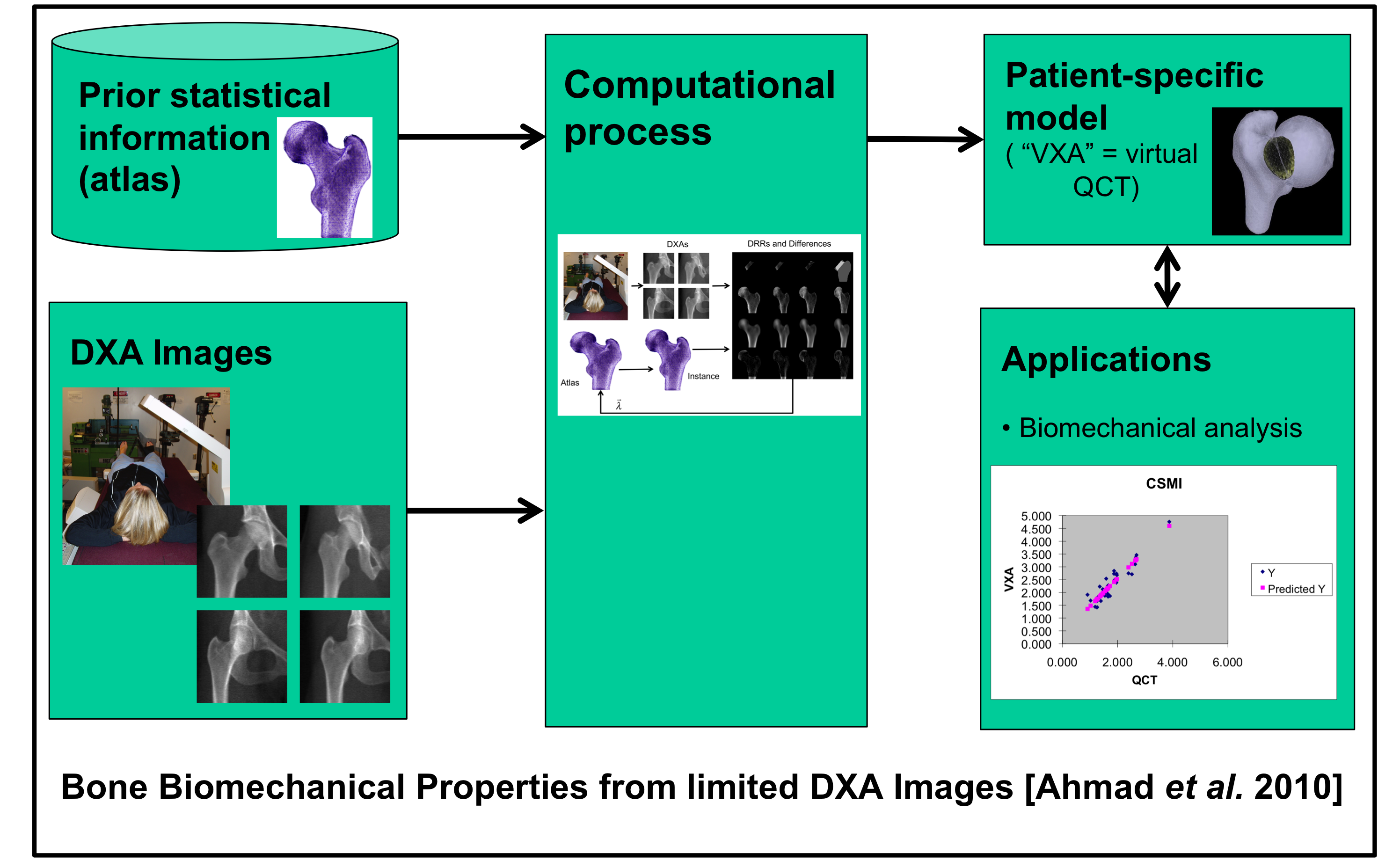 Another project has involved the use of a statistical atlas constructed from Quantitative CT (QCT)
images of the proximal femur [Wilson 2007] to construct “volumetric DXA (VXA)” models from a sparse set of
dual-energy (DXA) images [Ahmad 2010]. These VXA models are then used to compute biomechanical
measures that may be used to predict fracture risk.
Another project has involved the use of a statistical atlas constructed from Quantitative CT (QCT)
images of the proximal femur [Wilson 2007] to construct “volumetric DXA (VXA)” models from a sparse set of
dual-energy (DXA) images [Ahmad 2010]. These VXA models are then used to compute biomechanical
measures that may be used to predict fracture risk.
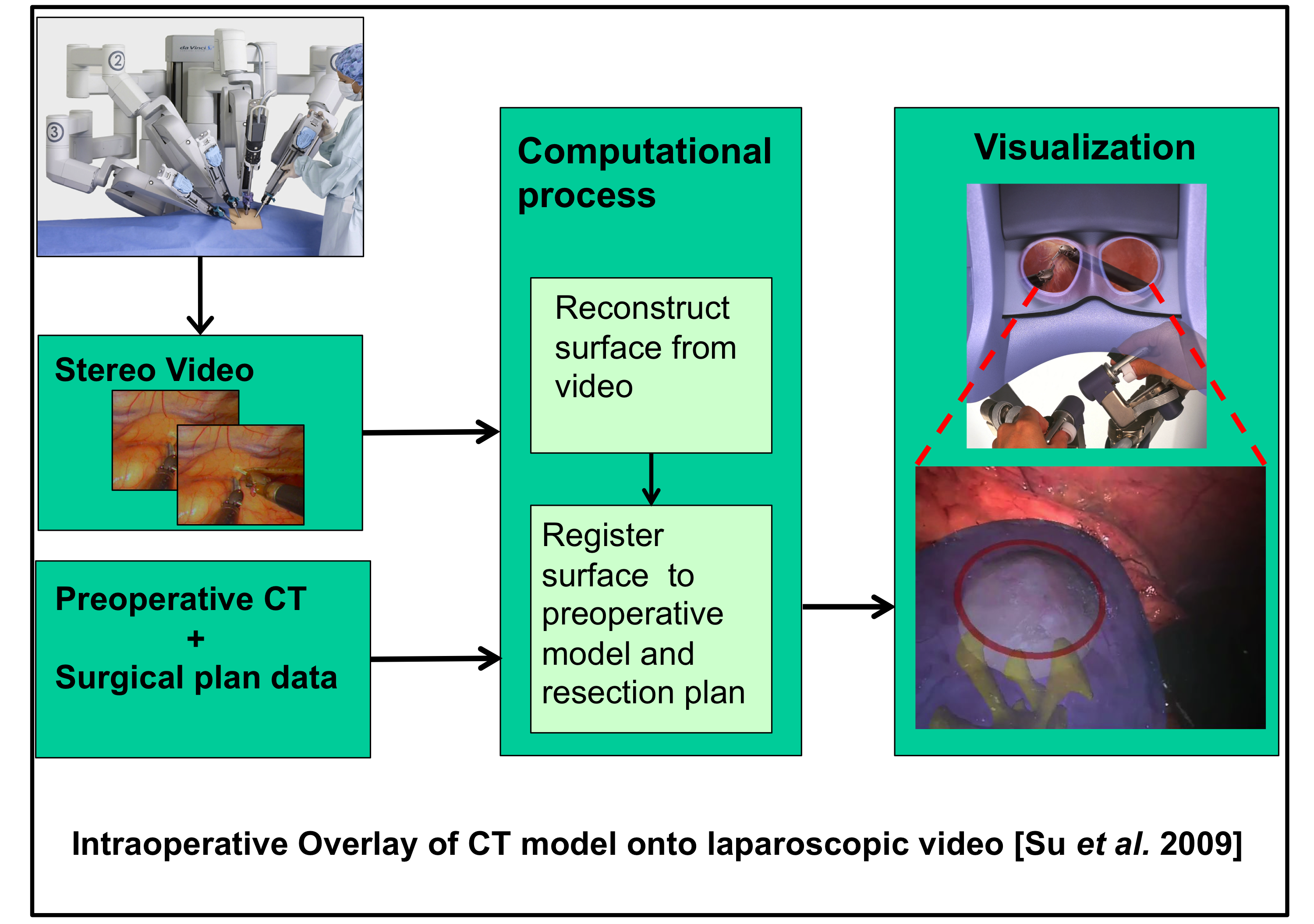 Other projects have emphasized the fusion of segmented preoperative models
with intraoperative endoscopic video. In one example [Su 2009],video
from a DaVinci stereo endoscope was used to create a 3D surface model
of a kidney undergoing laparoscopic partial nephrectomy. This model
was then registered to a segmented preoperative CT model to create
stereoscopic video overlay visualizations. In other projects
(e.g., [Burschka 2005], [Mirota 2009], [Mirota 2011], [Uneri 2012]), video
has been fused with segmented volumetric models for skull base and head and neck surgery.
Other projects have emphasized the fusion of segmented preoperative models
with intraoperative endoscopic video. In one example [Su 2009],video
from a DaVinci stereo endoscope was used to create a 3D surface model
of a kidney undergoing laparoscopic partial nephrectomy. This model
was then registered to a segmented preoperative CT model to create
stereoscopic video overlay visualizations. In other projects
(e.g., [Burschka 2005], [Mirota 2009], [Mirota 2011], [Uneri 2012]), video
has been fused with segmented volumetric models for skull base and head and neck surgery.
![]() The current application of interest is Total Face Transplant Surgery. The primary intent of this project is to estimate missing anatomical structure using a statistical shape model (SSM). This may be applied in the presence of an incomplete medical image or when a patient has undergone severe trauma. With an estimate of the patient's true anatomy, we believe that a more complete and accurate surgical plan may be used. A known, and non-traumatized, portion of the transplant candidate's cranial structure is matched to a SSM of the entire cranial structure. The regions instantiated from the SSM corresponding to unknown, or traumatized, regions of the patient's anatomy are used to estimate the entire cranial structure of the patient. The major issue with the completion approach described in [Chintalapani 2010] is the presence of a non-smooth transition from the known region to the unknown region. We hoped that by modeling this estimation problem as a Multivariate Gaussian Regression problem would yield a smooth boundary, however this was not the case. We believe that asymmetries resulting from the pose of the patient's mandible are not accurately modeled by the primarily symmetrical “known” region of the neurocranium used as prior data. In order to avoid this problem, work is currently underway to create separate SSMs for the neurocranium and mandible.
[
The current application of interest is Total Face Transplant Surgery. The primary intent of this project is to estimate missing anatomical structure using a statistical shape model (SSM). This may be applied in the presence of an incomplete medical image or when a patient has undergone severe trauma. With an estimate of the patient's true anatomy, we believe that a more complete and accurate surgical plan may be used. A known, and non-traumatized, portion of the transplant candidate's cranial structure is matched to a SSM of the entire cranial structure. The regions instantiated from the SSM corresponding to unknown, or traumatized, regions of the patient's anatomy are used to estimate the entire cranial structure of the patient. The major issue with the completion approach described in [Chintalapani 2010] is the presence of a non-smooth transition from the known region to the unknown region. We hoped that by modeling this estimation problem as a Multivariate Gaussian Regression problem would yield a smooth boundary, however this was not the case. We believe that asymmetries resulting from the pose of the patient's mandible are not accurately modeled by the primarily symmetrical “known” region of the neurocranium used as prior data. In order to avoid this problem, work is currently underway to create separate SSMs for the neurocranium and mandible.
[ ]
]
Details are currently on the intranet page.