Contact Us
CiiS Lab
Johns Hopkins University
112 Hackerman Hall
3400 N. Charles Street
Baltimore, MD 21218
Directions
Lab Director
Russell Taylor
127 Hackerman Hall
rht@jhu.edu
Last updated: Apr 27, 10:49 PM
The project is aimed at providing steady ultrasound imaging for needle biopsy. The main problem needs to be solved is to find proper feedback to control the robot. To solve this problem, B-mode images can be used a visual feedback. The transformation error can be estimated based on two ultrasound images, and then be used to complete the control loop.
Background
Tumor biopsy is the procedure to extract sample tissues or cells from the lesion or mass for diagnosis. And ultrasound imaging is used to visualize the lesion and guide the needle pass. During the procedure, there is a well established session to have pathologists in room, who can immediately check whether the acquired samples are diagnosable. Usually, several samples need to be required for biopsy.Therefore, the sonographers must hold the probe at the same position throughout the whole procedure, which is indeed cumbersome and tedious. And they have only one hand that can freely manipulate the needle.
Objectives
The aim of the project is to develop a robot-assisted system to navigate the probe and provide a steady view for the sonographers. There will be small motions due to organ slippage or breath. Based on the implement of deep learning, we would let the system estimate this distance(from current location to the target slice). And this information will be used to servo the robot.
Significance
The system will make repeatable biopsy more efficient and less cumbersome for the sonographers. Both of their hands are freed when the robot is there to help!
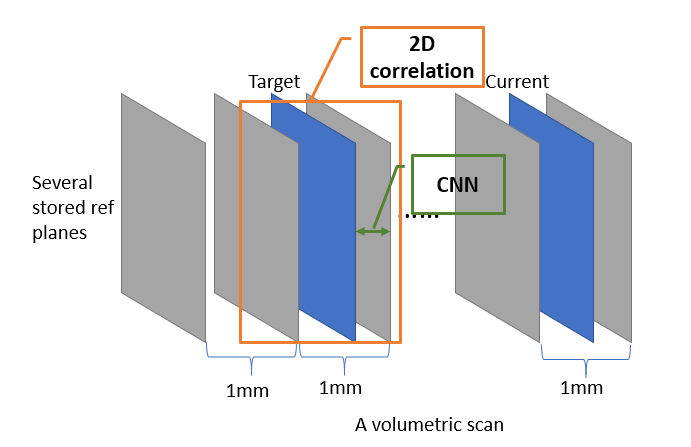
Out-of-plane motion: deep learning based on speckle decorrelation
The correlation of speckles in two images is a good estimation for elevational translation in ultrasound images. We would like to use this feature to train the neural network to perform an end-to-end learning.
First, a volume scan is obtained with a step size around 1 mm. Second, use the correlation of two images to find a large motion, bounding the target image (same for the current image) within two neighboring reference planes. Then use the CNN to estimate translation within 1 mm.
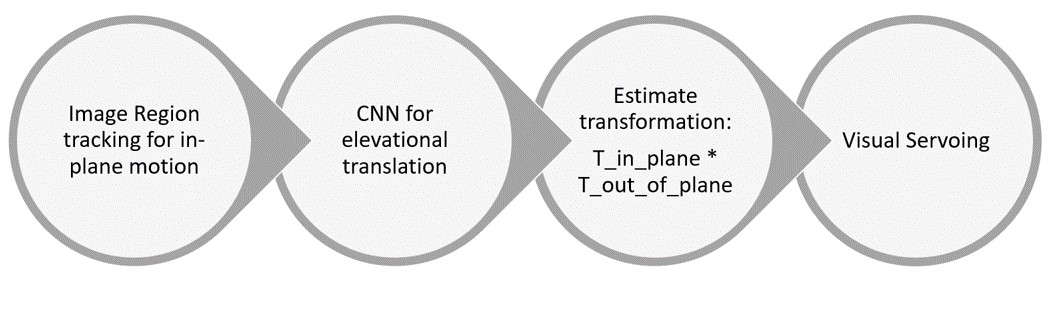
In-plane motion: Image region tracking algorithm
Following Hager and Belhumeur algorithem (1998).
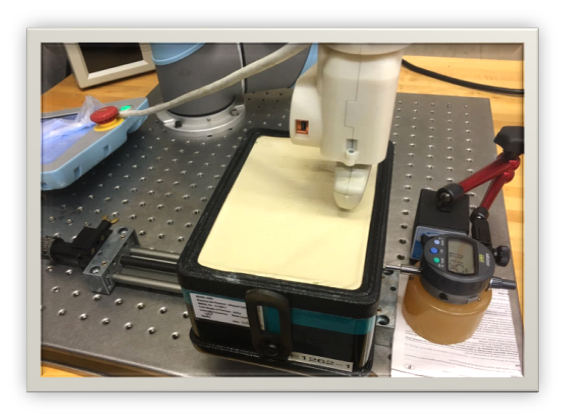
Testbed setup
Equipment: UR5, linear stage, dial indicator, CIRS elasticity phantom and Ultrasonix system.
Data: Image + corresponding position in the probe frame. Resolution: 0.05 mm. Range: 2 cm or +/- 2 degree.
describe dependencies and effect on milestones and deliverables if not met
UR5: Resolved
Phantoms: Resolved
Computation power: Resolved
Linear stage: Resolved
Dial indicator: Resolved
Access to 3D printer: Resolved
}
- A. A. Azar, H. Rivaz and E. Boctor, “Speckle detection in ultrasonic images using unsupervised clustering techniques,” 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, 2011, pp. 8098-8101.
- Hager, G., & Belhumeur, P. N. (1998). Efficient region tracking with parametric models of geometry and illumination. IEEE Transactions on Pattern Analysis and Machine Intelligence, 20(10), 1025-1039. https://doi.org/10.1109/34.722606
- Laporte, C., & Arbel, T. (2011). Learning to estimate out-of-plane motion in ultrasound imagery of real tissue. Medical Image Analysis 15, 202-213. doi:doi.org/10.1016/j.media.2010.08.006
- Prager R.W., Rohling R.N., Gee A.H., et al (1998). Rapid calibration for 3-D freehand ultrasound. Ultrasound Med Biol 1998b;24:855-869
- Prager R.W.,Gee A.H., Treece G.M., Berman L.H.(2003).Decompression and speckle detection for ultrasound
images using the homodyned k-distribution.Pattern Recognition Letters, Volume 24, Issues 4–5,2003,Pages 705-713,ISSN 0167-8655, https://doi.org/10.1016/S0167-8655(02)00176-9.
- Salehi, R. M., Sprung, J., Bauer, R. & Wein, W. (2017). Deep Learning for Sensorless 3D Freehand Ultrasound Imaging. In: Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. MICCAI 2017.
======Other Resources and Project Files======
Here give list of other project files (e.g., source code) associated with the project. If these are online give a link to an appropriate external repository or to uploaded media files under this name space (2019-99).